Research Article - Biomedical Research (2018) Volume 29, Issue 17
Antibiotic susceptibility and plasmid profile of clinical isolates of Escherichia coli
Badger-Emeka Lorina Ineta1*, Emeka Promise Madu2, Al-Sultan Abdulrahman Abdulhadi1, Hairul Islam M Ibrahim31Microbiology Division, Department of Biomedical Sciences, College of Medicine, King Faisal University, 31982, Hofuf, Saudi Arabia
2Department of Pharmaceutical Science, College of Clinical Pharmacy, King Faisal University, 31982, Hofuf, Saudi Arabia
3Department of Biological Sciences, College of Science, King Faisal University, 31982, Hofuf, Saudi Arabia
- *Corresponding Author:
- Badger-Emeka Lorina Ineta
Microbiology Division
Department of Biomedical Sciences
College of Medicine, King Faisal University
Saudi Arabia
Accepted date: September 09, 2018
DOI: 10.4066/biomedicalresearch.29-18-927
Visit for more related articles at Biomedical ResearchAbstract
Background/Aims: The role of mobile genetic elements such as plasmids in the spread of multiple antibiotic resistance has continued to receive attention by researchers. Due to the diverse nature of Escherichia coli strains characteristically harbouring large proportion of genomic plasmids, the aim of the present study is to evaluate the antimicrobial susceptibility and plasmid profiles of clinical isolates of E. coli in this south eastern region of Saudi Arabia.
Methods: Bacterial isolation, antimicrobial susceptibility test were carried out using basic microbiological techniques. Dendrogram of clustering in relationship to antibiotic susceptibility of the isolates was carried out using PAST. Extracted plasmid DNA was visualised using the ultraviolet light illuminator and photo documentation system. Sizes of Plasmid were determined using standard DNA molecular weight markers at 1kb ladder. Plasmid curing was carried out using Sodium Dodecyl Sulphate (SDS).
Results and Conclusion: Antibiotic resistance ranged between 85.71%-100% for the cephalosporins, 64.29% to 92.9% resistance for penicillins, including Aztreonam, a last line drug. Results indicate that 70% of the isolates did not show any plasmid presence. However, 30% of the isolates contained plasmids with molecular weights of 30kb and above. Dendrogram of clustering in relationship to antibiotic susceptibility showed isolates were in 3 clades pre-plasmid curing and 6 clades post-plasmid curing. One isolate (EC36) was cured of its resistance to Penicillins, while isolates EC67, EC77 and EC87 initially sensitive to ciprofloxacin, became resistant to the antibiotic. The findings suggest a preponderance of chromosomal mediated resistant E. coli. The few plasmid containing resistant bacteria, were resistant to curing as they displayed continued antibiotic resistance after curing. This therefore confirms the growing presence of multiple genomic plasmid harbouring E. coli.
Keywords
Multi-antibiotic resistance, Bacterial infections, Plasmids, E. coli, Isolates.
Introduction
The exhibition of multi-antibiotic resistance by bacterial in recent times continues to constitute a major public health problem that is yet to be resolved. According to a 2013 report by Centers for Disease Control and Prevention [1], contributory factors to the spread of antibiotic resistance in bacteria is poorly understood. Thus this rapid rise in difficult to treat bacterial infections has been described by public health organisations as a potential crisis leading to subsequent “catastrophic consequences” [2]. In a recent write up by Joshi [3], it is postulated that by the year 2050, there would be an estimated 10 million people dying on yearly basis as a result of difficult to treat bacterial infections. The write up further points to the fact that “a major new class of antibiotic was discovered 30 years ago”; thus, indicating that antibiotics are not being produced as fast as they are needed. Earlier, a CDC [1] report declared the human race to be in a “post anti-biotic era”, with the World Health Organisation (WHO) warning that resistance to antibiotics has become dire [4]. This is particularly the case with gram Negative bacteria pathogens which are resistant to most of the antibiotic options available, thus creating scenarios similar to the pre-antibiotic era [1,5]. One of such gram negatives is Escherichia coli which was listed to be among the cause of serious infections even in community settings with an increasing prevalence including extended-spectrum betalactamase producing E. coli [5].
Escherichia coli is generally a large and diverse bacterium that colonises both human and animal intestine as well as being found in the environment. This bacterium ranges from being harmless to pathogenic, a leading cause of urinary tract infections, respiratory infections [6-8], pneumonia [9], global food poisoning [10-12]. In all cases of pathogenic E. coli infections resistance to drugs of choice have been previously reported [13] with isolates exhibiting multi-drug resistance (MDR) a view that had also been expressed in an earlier report [14].
There is a wide spread of resistance to antimicrobials by infections resulting from E. coli and this is receiving much attention globally, particularly so with the advent of multiantibiotic resistant plasmid genes with the example of Extended spectrum β-lactamases (ESBLs) which is plasmid mediated. In other cases, bacteria have been reported to be resistant to colistin, an antibiotic considered as a last line one. Resistance genes to this last line antibiotic has been located not on the bacteria chromosomal DNA but on the extrachromosomal plasmid DNA. Generally plasmids have been reported to spread genes responsible for resistance to antibiotics and according to Savara and Rankin [15] when the resistance genes are mobile, carried on plasmids, the results are acceleration in their spread. A wide range of plasmids carrying MDR genes constitutes a major medical problem as these genes are easily transmissible to other bacteria species [16,17]. The results in rapid and wide spread of plasmid mediated antibiotic resistance is a global occurrence as with the example seen in colistin resistance [18,19].
Two genetic bacteria systems, chromosomal DNA and extrachromosomal DNA, are contributory to the transfer of genes through many processes involving mutations, recombination and horizontal transfer of genes with a resultant bacterial genomic diversity. Thus, MDR-plasmids help spread resistance by horizontal gene transfer [20]. Strains of Escherichia coli are generally reported to have a large proportion of genomic plasmid codes responsible for resistance to antibiotics than those of the chromosome [15]. The fact that antibiotics are not being produce to meet up with the rate at which bacteria resistant strains are evolving, there is the postulation that biotechnology might be a useful tool needed to cub the transfer of resistant genes. To this effect, Kamruzzaman et al. [21] demonstrated that plasmid interference could be used in creating plasmid incompatibility as a means for curing of antibiotic resistance plasmid genes. They recommended that plasmid research may be a useful tool suitable for clinical use in individuals who are found to have been colonised. Earlier, van Hal et al. [22] had demonstrated that the eradication of plasmids detected in the gut flora of patients was capable of preventing further complications such as sepsis that would have resulted from resistance to antibiotics in patients. It is therefore pertinent that there be a continual monitoring of resistance to antimicrobials by bacteria isolates as well as characterising the genes responsible for such resistance. Due to the fact that the region is characterised by high antimicrobial resistance, and because of the diverse nature of E. coli, the findings would help in highlighting whether resistance as encountered in the isolates are chromosomally or extra chromosomally mediated. The present investigation looks to evaluate the antimicrobial susceptibility and plasmid profile of MDR clinical isolates of Escherichia coli from hospitalised patients in Al-Ahsa, south eastern region of Saudi Arabia.
Materials and Method
Ethical consideration
The E. coli isolates used for the study were obtained as part of routine patient care, no patient consent was required while the research was approved by the research and ethics committee of college of Medicine, King Faisal University.
Isolation of E. coli and antimicrobial assay
Samples had been collected from hospitalised patients in ICU, OPD, ER and Wards. Source of isolation included blood, bedsores, catheters, placenta, urine, eye swaps, wound swaps, peritoneal fluid and cervical swaps. Collected samples were plated out on MacConkey agar and cultured aerobically at 37°C for 24 h. Basic bacteriological technique was used for the isolation of E. coli while confirmation of the obtained isolates were carried out using the Vitek 2 compact automated system (BioMerieux, Marcy L’Etoile, France) according to the manufacturers guidelines, using GN cards. Antimicrobial susceptibility was carried out using the AST cards with the following antibiotics: Ampicillin, Ampicillin/Sulbactam, Piperacillin, Piperacillin/Tazobactam, Cefazolin, Cefuroxime, Cefuroxime Axetil, Cefoxitin, Cefpodoxime, Cefotaxime, Ceftazidime, Cefepime, Imipenem, Meropenem, Gentamicin, Tobramycin, Ciprofloxacin, Levofloxacin, Tigecycline, and Sulphamethoxazole/Trimethoprim.
A total of confirmed 50 E. coli isolates exhibiting MAR to the antibiotics were subjected to plasmid extraction.
Plasmid extraction and analysis
Pure E. coli isolates were used for the extraction of plasmid DNA using the Carolina biological mini prep kit according to manufacturers’ guidelines. The extracted plasmids were processed by electrophoresis on 1% agarose gel and stained with ethidium bromide. The visualisation of obtained plasmid DNA was carried out using the ultraviolet light illuminator and photo documentation system used for analysis. Plasmid sizes were determined using standard DNA molecular weight markers at 1 kb ladder.
Plasmid curing of isolates
Fifteen of the E. coli isolates with plasmids were subjected to curing using the method of Zaman et al. [23] but with modifications. Each of the test organisms was culture in a solution of Nutrient broth containing ampicillin for 24 h at a temperature of 37°C, aerobically. The overnight growth was diluted to 104 cell/mL with nutrient broth using the spectrophotometer (BOECO Germany). The optical density was determined using an online E. coli cell culture concentration from OD600 calculator. The appropriate cell concentrations for each of the 15 E. coli samples were diluted to 104 cells/ml in nutrient broth. For each of the sample, a 0.5 mL of the obtained dilution was added to 4.5 mL of nutrient broth containing 10% of Sodium Dodecyl Sulphate (SDS), bringing the concentration to 103 cells/mL. The resultant was incubated aerobically at 37°C in an orbital shaker for 48 hours. The post 48 hour’s growth was diluted with normal saline to obtain a 103 cells/ml. 1ml of the resultant for each of the isolate was inoculated and spread out on Muller Hilton agar and incubated aerobically at 37°C for 24 h.
Antimicrobial susceptibility post- plasmid curing
The disk diffusion method of Bauer and Kirby was used for the antimicrobial assay of post plasmid curing experiment of the isolates. Obtained colonies were selected and plated out on Muller Hilton agar containing the following antibiotic disks: ampicillin (10μ), ampicillin/sulbactam (10μ), cefotaxime and ceftazidime 30μ each, ciprofloxacin 5μ and amikacin 30μ. All plates were incubated aerobically overnight at 37°C. The zones of inhibition were measured (mm) using a millimetre rule and the results were compared with the zones of inhibition for the initial zones of inhibition for the clinical E. coli isolates. The recommended standards by National Committee for Clinical Laboratory Standards for the interpretation of zone of inhibition were used for the interpretation of zones of inhibition [24].
Phylogenetic grouping based on antimicrobial susceptibility
The phylogenetic relatedness of 45 of the E. coli isolates based on the resistance profile were generated with the “unweighted pair group average (UPGMA) algorithms” of PAST.
Statistical analysis
Isolates were classified as resistant, intermediate and sensitive using the CLSI 2015 guide for the interpretation of zones of inhibition. Antimicrobial resistance were determined in percentages.
Results
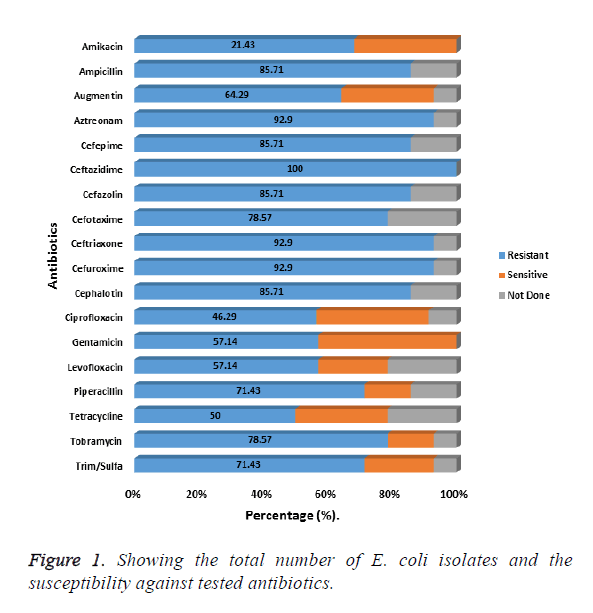
Fifty E. coli isolates exhibiting MDR against the tested antibiotic were used for the study. Most of the isolates (38%) had been isolated from wounds and urine samples (24%). For the results on antibiotic susceptibility of the 50 E. coli isolates, the results presented in Figure 1 shows resistance to the following, cephalosporin’s, ceftazidime, cefuroxime, ceftriaxone to be, 100%, 92.9% and 92.9% respectively. For other cepharlosporins, like cephalotin, Cefepime, cefazolin, each showed an 85.71% resistance each. The least percentage resistance for the Cepharlosporins was 78.57% encountered with cefotaxime (Figure 1).
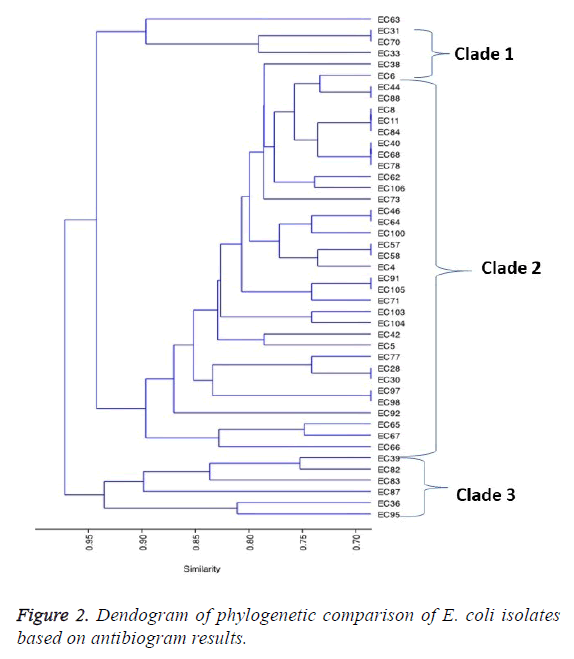
For the β-lactams, resistance for the following antibiotics Aztreonam, ampicillin, piperacillin and Augmentin were as follows: 92.9%, 85.71%, 71.43% and 64.29% respectively. Of the fifty isolates, the phylogenetic relatedness of 45 of those isolates that showed extreme drug resistance (XDR) was used for the comparison. The resulting dendrogram on the phylogenetic relatedness based on the isolates susceptibility to antimicrobials is shown in Figure 2. Clustering by UPDMA algorithm determined on Jaccard shows three clades consistent with the isolates based on the similarity coefficients for all the isolates used in the study. The prevalent resistant E. coli were distributed in clade 2, constituting 75.6% of the isolates. In clades 1 and 3, were 6.7% and 13.3% of the isolates respectively. The remaining 2 (4.4%) of the isolates were not in the three main clades (Figure 2).
Plasmid extraction
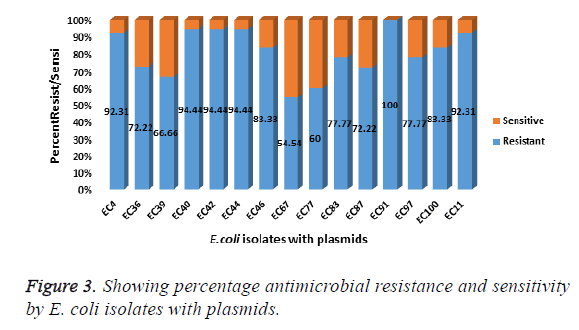
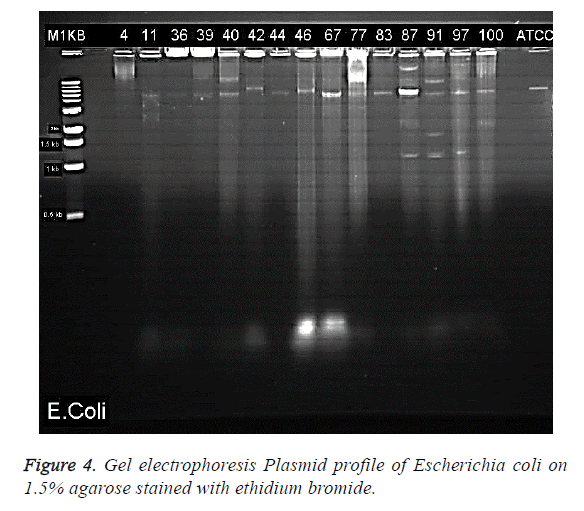
Gel electrophoresis results of the visualised plasmid DNA are showed that fifteen of the isolates (30%) had plasmids extracted while the majority of 70% were without plasmids (Table 1). The percentage resistance for each of the 15 isolates with plasmid were as follows: EC4 (92.31%), EC11 (92.31%), EC36 (72.22%), EC39 (66.66%), EC40 (94.44%), EC44 (94.44%), EC46 (83.33%), EC67 (54.54%), EC77 (60%), EC83 (77.77%), EC87 (72.22%), EC91 (100%), EC97 (77.77%), EC100 (83.33%) (Figure 3). The molecular weights of the plasmid DNA of these E. coli isolates varied in sizes (Figure 4). The figure shows that for all the isolates, the number of bands per plasmid varied, ranging from one to four with different molecular weights (MW). All the fifteen E. coli contained plasmids with 30 kb and above. Nine of the isolates showed only one band size and this range from 30kb and above (Table 3). The remaining isolates had plasmids with different number of bands. The following isolates EC40, EC77, EC97 and EC100 showed 2 bands which ranged from above 35 kb to 15 kb. The plasmid of one isolate, EC87 showed 3 bands with MW ranging between 10 kb to 70 kb. The most number of bands were seen in isolate EC91 which showed a total of four bands that ranged from 10 kb to above 60 kb (Table 3). It is also worth noting that this isolate (EC91) exhibited a 100% resistance to the tested antibiotics.
| Source of Isolation | Total Number | Plasmid | None |
|---|---|---|---|
| Blood | 4 (8%) | 2 | 2 |
| Urine | 12 (24%) | 3 | 9 |
| Wound swap | 19 (38%) | 7 | 12 |
| Trach Asp | 1 (2%) | 0 | 1 |
| Eye swab | 1 (2%) | 0 | 1 |
| Cervical WD | 2 (4%) | 1 | 1 |
| peritoneal fluid | 3 (6%) | 0 | 3 |
| Cath Tip | 2 (4%) | 1 | 1 |
| drain tip | 3 (6%) | 1 | 2 |
| Skin Swaps | 1 (2%0 | 0 | 1 |
| placenta | 1 (2%) | 0 | 1 |
| bedsore | 1 (2%) | 0 | 1 |
| Total | 50 (100%) | 15 (30%) | 35 (70%) |
Table 1: Showing source of E. coli isolation and presence or absence of plasmids.
| E. coli isolates | Antibiotics | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | Ampicillin/Sulbactam | Cefotaxime | Ceftazidime | Ciprofloxacin | Amikacin | |||||||||||||
| B/F | A/T | I | B/F | A/T | I | B/F | A/T | I | B/F | A/T | I | B/F | A/T | I | B/F | A/T | I | |
| EC4 | 4 | 2 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 7 | 10† | R | 2 | 2 | R |
| EC11 | 2 | 2 | R | 0 | 0 | R | 0 | 2 | R | 0 | 2 | R | 8 | 10† | R | 0 | 0 | R |
| EC36 | 2 | 20* | S | 0 | 2 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 22* | 25† | S |
| EC39 | 2 | 10† | R | 0 | 2 | R | 0 | 0 | R | 0 | 0 | R | 2 | 6† | R | 17 | 17 | S |
| EC40 | 0 | 2 | R | 0 | 2 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 18 | 19† | S |
| EC42 | 0 | 2 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 15 | 14 | I |
| EC44 | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 14 | 14 | I |
| EC46 | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 15 | 15 | I |
| EC67 | 0 | 0 | R | 0 | 2 | R | 0 | 0 | R | 0 | 0 | R | 20 | 8** | R | 18 | 16 | I |
| EC77 | 0 | 5† | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 20 | 10** | R | 20 | 19 | S |
| EC83 | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R |
| EC87 | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 15 | 6** | R | 18 | 17 | S |
| EC91 | 0 | 2 | R | 0 | 2 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 20 | 19 | S |
| EC97 | 0 | 3† | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 17 | 17 | S |
| EC100 | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 0 | 0 | R | 18 | 18 | S |
Table 2: Showing the zones of inhibition by E. coli isolates before and after plasmid curing experiments.
| Escherichia coli strain | Percentage Resistance (%) | Number of Bands | Plasmid Product (Kb) | Clade group | |
|---|---|---|---|---|---|
| B | A | ||||
| EC4 | 92.31 | 1 | <50 | 2 | 0 |
| EC11 | 92.31 | 1 | 50 | 2 | 4 |
| EC36 | 72.22 | 1 | 50 | 3 | 2 |
| EC39 | 67 | 1 | 50 | 3 | 2 |
| EC40 | 94.44 | 2 | 50, 70 | 2 | 3 |
| EC42 | 94.44 | 1 | 60 | 2 | 3 |
| EC44 | 94.44 | 1 | 50 | 2 | 4 |
| EC46 | 83.33 | 1 | 50 | 2 | 0 |
| EC67 | 55 | 1 | 50 | 2 | 5 |
| EC77 | 60 | 2 | 50, 60 | 2 | 6 |
| EC83 | 78 | 1 | 50 | 3 | 1 |
| EC87 | 72.22 | 3 | 70, 50, 10 | 3 | 1 |
| EC91 | 100 | 4 | 60, 50, 15, 10 | 2 | 0 |
| EC97 | 78 | 2 | 50, 10 | 2 | 5 |
| EC100 | 83 | 2 | 50, 20 | 2 | 6 |
Table 3: Showing relationship between antibiotic resistance and number of plasmid bands and sizes.
Post plasmid curing
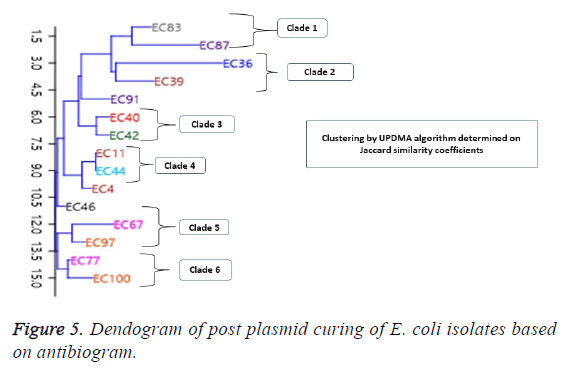
The post plasmid curing antibiotic susceptibility results are shown in Table 2. Selected for the test were antibiotics in the groups of β-lactams, fluoroquinolones and aminoglycosides. Results showed majority of the isolates remained resistant against the tested drugs. For the penicillins, one isolate EC36, was cured and became sensitive to the ampicillin but not to Augmentin antibiotic as well as retaining it sensitivity to Amikacin. For other isolates such as EC39, EC77 and EC97, though the isolates remained resistant against ampicillin, these isolates exhibited a wider zone of inhibition after plasmid curing. However, all the 15 E. coli isolates remained resistant to the cephalosporins post curing experiments. For the fluoroquinolones, all isolates remained resistant. However, EC4 and EC11, showed wider zones of inhibition after curing as shown in the Table 2. A different scenario was encountered for three other isolates, EC67, EC77 and EC87 that were initially sensitive to the fluoroquinolone antibiotic became resistant after plasmid curing. The results also showed that all the E. coli isolates which were sensitive to amikacin remained sensitive, however, for some the zones of inhibition remain unchanged (EC39, EC44, EC97, EC100). For others, EC36, EC40, there was an increased zone of inhibition while in the remaining isolates, there was a slight decrease in zones of inhibition but isolates remained sensitive (Table 2). In terms of the post plasmid curing phylogenetic relatedness based on antibiotic susceptibility of the E. coli isolates, the dendrogram of clustering by UPGMA algorithm based on Jaccard similarity coefficient shows an even distribution of the isolates into 6 clades (Figure 5). The distribution is consistent in some cases, with the number plasmid bands of the isolates in each clade. In clade 1, EC83 and EC87 had different bands in the plasmid while the isolates in clade 2 each showed one band greater than 50kb. In the third clade consisting of EC40 and EC42 showing one and two bands respectively, both of which range between 50 kb and 70 kb. A similar pattern is exhibited by other isolates in clade 4 and 5. Escherichia coli isolate EC91, EC4 and EC46 showed no phylogenetic relationship with the other groups (Figure 5).
Discussion
The results from this study show high antimicrobial resistance encountered among the E. coli isolates. This resistance had cut across all the isolates regardless of the source as well as the various classes of antibiotics tested.
Worthy of note is that the isolates can also be grouped as ESBL producing pathogens on the basis of the results on antimicrobial susceptibility test. The high antimicrobial resistance by E. coli had previously been reported by researchers [13,25] with Alsultan et al. [25] reporting the antibiotic susceptibility by ESBL producing E. coli from the region of the present study. All the fifteen E. coli isolates with plasmids had the characteristics to be described as ESBL producing as they had been resistant to the penicillins and cephalosporins. It would therefore have been expected that since resistance by ESBLs producers is plasmid mediated plasmid curing would have resulted in a more sensitivity to the antibiotics against which they had been initially resistant. However, only one isolate (EC36) was seen to have lost its resistance to penicillin while retaining its’ resistance to other antibiotics but remained sensitive to aminoglycoside. There are a number of possible contributory factors to the obtained results. Plasmid curing is said to occur naturally by various means one of which is through the chemical treatment of cells [26]. The effectiveness of curing agents has been reported to vary and that this variation could range from 100-1000 fold [27] also, that the variations could depend on the efficacy of the curing agent and the organism being cured. Sodium Dodecyl Sulphate (SDS) was used for plasmid curing in the present study according to Zaman et al. [23] which Inuzuka et al. [28] had earlier recommended suitable for plasmid curing among E. coli isolates. We report that it might have been a contributory factor to the resistance seen post plasmid curing of the E. coli isolates as seen in the obtained results. This is due to the fact that effectiveness of curing agents vary with earlier reports by Aja et al. [29] indicating that 10% of SDS did not cure the isolates in their study. On the other hand, Schaufter et al. [30] found and reported that Ethidium bromide and Acridine orange were not effective in the curing of ESBLplasmid curing strains in their study, with a similar observation reported from an earlier study [31]. Thus SDS was chosen for the present study according to available literature, bearing in mind that the effectiveness of curing agents varied with different potential weaknesses [27].
Besides the curing agent used, the obtained results could be attributed to other possible reasons. The gel electrophoresis results showed the molecular weight of the most of the isolates to have ranged between 30kb and above. The number and size of the plasmids in the tested isolates varied significantly in the present investigation, ranging 10kb at above 50kb. These findings are similar to those from other regions of the world. Earlier, Nasreen et al. [32] while analysing the plasmid profile of E. coli from urine samples reported the presence of single sized as well as multiple sizes among their isolates. They observed a high antibiotic resistance among isolates that had molecular weight of 23 kb-26 kb. They reported a high MDR among all their isolates as well as finding that about 50% of the isolates possessed plasmids. In the present study plasmids were encountered in only 30% of the isolates despite the fact that all the isolate were MDR with features of being ESBL producers. With high resistance being attributed to plasmids having molecular weights of 23-26 kb, the resultant high resistance as seen in this study could be due to the 30kb and above of the molecular weight of the plasmids as well as the presence of multiple sizes amongst the isolates. However, it is known that E. coli isolates carry multiple plasmids which could be the reason why they are resistant to antibiotics [33]. The only isolate in the present investigation with 100% resistance against the tested antibiotics (EC91) was seen to carry multiple bands with different sizes and this could be the contributory factor in the exhibited resistance in this isolate. Also studies have shown that penicillin resistant E. coli harboured plasmids that are within the range of 30 kb and above [33], such as is the case of the E. coli isolates in the present investigation. Thus, E. coli showing MDR have been reported to carry multiple plasmids of different sizes and this has been the case of those encountered in this study.
Remarkably of the 50 MDR isolates used for this investigation, there were only 15 with plasmids suggesting that the resistance in the other isolates was probably chromosomally mediated. Also, resistance can be generally grouped as either being chromosomal when cells have not been affected by curing of plasmids or could be “plasmodial when cured [27]. Of the 15 isolates subjected to plasmid curing; only one of the isolates (EC36) was cured and became sensitive to penicillin but remained resistant to the cephalosporins. It could possibly mean that resistance was both plasmid mediated as well as chromosomally mediated in these clinical isolates of E. coli from the region of the present study. Reboucas et al. [34] demonstrated that isolates in their study remained resistant to ampicillin after curing of plasmids and suggested that resistance could have been mediated chromosomally. Similar findings have also been reported in more recent researches [35].
Also worthy of note is the loss of sensitivity by some of the isolates (EC67, EC77 and EC87) to ciprofloxacin, an antibiotic they had been initially sensitive to. The results though puzzling can be attributed to a number of possible factors. One of which might be the plasmid curing method. It has been suggested [36] that the curing methods being used were capable of inducing mutations in the host DNA. It would therefore be expected that curing agents would produce different effects according to bacteria species. The fact that curing agents had been suggested to cause possible mutations in host chromosomes and thereby interfering with plasmid analysis [27] might explain the reason for the loss of sensitivity to ciprofloxacin as seen in the present study. Also the initial phylogenetic comparison of the clinical isolates base on their resistance to antimicrobials showed close relatedness with majority of the isolates showing cluster similarity in clade 2. This similarity was independent of the source of isolation. Similar findings had been reported by Carvalho et al. [37] who found that the most prevalent phylogenetic group was irrespective of the source of isolates. The antimicrobial susceptibility of the isolates post plasmid curing showed further variation in phylogenetic grouping, resulting in 6 clades. This suggests the possible presence of additional factors which could be acting together to create variability. The sizes and band numbers might be contributory reasons for this variability. Multiple reasons have been postulated as being responsible for the presence of multiple bands with varying sizes in plasmid DNA. However, the high versatility generally exhibited by E. coli isolates has been characterised on a number of variables in terms of their resistance to antimicrobials by researchers [38]. Also, variability according to Casellas et al. [39] could be influenced by the spread of plasmids as well as other mobile carriers of resistant genes.
The problem of resistance by bacteria to antibiotics continues and if as suggested by Kamrzzaman et al. [21], hospitals managing critically ill patients who are at high risk of being infected, there might need to look into the possibility of using simple oral administrative route of in vivo plasmid curing. This could be the much need light to the threat of antibiotic resistance.
It can therefore be concluded that the majority of the multidrug resistant E. coli isolates used in this study, did not have plasmids. However, in cases with positive plasmids, they were seen to have high molecular weights and an exhibited high resistant to antibiotics. Also, some of the isolates that were cured of plasmids, continued to be resistant to multiple antibiotics suggesting the possibility that they are carrying multiple resistance gene. This also portends danger as these genetic materials can be transferred to not only other E. coli strains but to other bacterial pathogens. The findings still highlights the presence of very unusual bacteria carrying multiple resistant gene that are within the region of the present study.
Acknowledgement
The researchers would like to thank Mr. Hani Al-Farhan, Mr. Hani Al-Rasisi, Ms Hajer Salman Al-Dehailan and Ms Fatimah Mohammed Said Al-Guaimy, for their technical assistance.
References
- Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention 2013.
- Viswanathan VK. Off-label abuse of antibiotics by bacteria. Gut Microbes 2014; 5: 3-4.
- Tina J. Antibiotic Resistance and Microwaves. Biomed Scientist 2018.
- Michael CA, Dominey-Howes D, Labbate M. The antibiotic resistance crisis: causes, consequences, and management. Front Public Health. 2014; 2: 145.
- Rossolini GM, Arena F, Pecile P, Pollini S. Update on the antibiotic resistance crisis. Curr Opin Pharmacol 2014; 18: 56-60.
- Shah MU, Mahmood M, Usman J, Kaleem F, Khalid Ali. Gram negative organisms in community acquired respiratory tract infections. J Microbiol Infect Dis 2013; 3: 8-11.
- Khan S, Singh P, Sachan A. Bacteria etiological agents causing lower respiratory tract infections and their resistance patterns. Iran Biomed J 2015; 19: 240-246.
- Prat C, Lacoma A. Bacteria in the respiratory tract-how to treat? Or do not treat? Int J Infect Dis 2016; 51:113-122.
- Mathilde PH, Jean-Damien R, Arnaud F, DeborahYH, Stéphane G, Julie B. Pathophysiology of Escherichia coli pneumonia: Respective contribution of pathogenicity islands to virulence. Int J Med Microbiol 2018; 308: 290-296.
- Al-Zogibi OG, Mohamed MI, Hessain AM, El-Jakee JK, Kabli SA. Molecular and serotyping characterization of shiga toxogenic Escherichia coli associated with food collected from Saudi Arabia. Saudi J Biol Sci 2015; 22: 438-442.
- Bandyopadhyay S, Lodh C, Rahaman H, Bhattacharya D, Bera AK. Characterization of Shiga toxin producing (STEC) and enteropathogenic Escherichia coli (EPEC) in raw yak (Poephagus grunniens) milk and milk products. Res Vet Sci 2012; 93: 604-610.
- Omar KB, Barnard TG. Detection of diarrhoeagenic Escherichia coli in clinical and environmental water sources in South Africa using single-step 11-gene m-PCR. World J Microbiol Biotechnol 2014; 30: 2663-2671.
- Alanazi MQ, Alqahtani FY, Aleanizy FS. An evaluation of E. coli in urinary tract infection in emergency department at KAMC in Riyadh, Saudi Arabia: retrospective study. Ann Clin Microbiol Antimicrob 2018; 17: 3.
- Khawcharoenporn T, Vasoo S, Singh K. Urinary tract infections due to multidrug-resistant enterobacteriaceae: prevalence and risk factors in a Chicago Emergency Department. Emerg Med Int 2013; 2013: 258517.
- Svara F, Rankin DJ. The evolution of plasmid-carried antibiotic resistance. BMC Evol Biol 2011; 11: 130.
- Mollenkopf DF, Stull JW, Mathys DA, Bowman AS, Feicht SM, Grooters SV. Carbapenemase-producing Enterobacteriaceae recovered from the environment of a swine farrow-to-finish operation in the United States. Antimicrob Agents Chemother 2017; 61: e01298-e013016.
- Ridenhour BJ, Metzger GA, France M, Gliniewicz K, Millstein J, Forney LJ, Top EM. Persistence of antibiotic resistance plasmids in bacterial biofilms. Evol Appl 2017; 10: 640-647.
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161-168.
- Poirel L, Kieffer N, Liassine N, Thanh D, Nordmann P. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. The Lancet Infect Dis 2016; 16: 281.
- Banu H, Prasad KP. Role of plasmids in microbiology. J Aquac Res Development 2017; 8: 466.
- Kamruzzaman M, Shoma S, Thomas CM, Partridge SR, Iredell JR. Plasmid interference for curing antibiotic resistance plasmids in vivo. PLoS ONE 2017; 12: e0172913.
- van Hal SJ, Wiklendt A, Espedido B, Ginn A, Iredell JR. Immediate appearance of plasmid-mediated resistance to multiple antibiotics upon antibiotic selection: an argument for systematic resistance epidemiology. J Clin Microbiol 2009; 47:2325-2327.
- Zaman MA, Pasha MH, Akhter MZ. Plasmid curing of escherichia coli cells with ethidium bromide, sodium dodecyl sulfate and acridine orange. BJM 2010; 27: 28-31.
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty Fifth Informational Supplement M100-S23; CLSI: Wayne, PA, USA, 2015.
- Alsultan AA, Aboulmagd E, Amin TT. ESBL-producing E. coli and K. pneumoniae in Al-Ahsa, Saudi Arabia: antibiotic susceptibility and prevalence of blaSHV and blaTEM. J Infect Dev Ctries 2013; 7: 1016-1019.
- Elias HM, Qader MK, Salih WM. Determination of plasmid DNA role in multidrug resistant Pseudomonas aeruginosa clinical isolates. Inter J Microb Immunol Res 2013; 1: 80-86.
- Letchumanan V, Chan K-G, Lee L-H. An insight of traditional plasmid curing in Vibrio species. Front Microbiol 2015; 6: 735.
- Inuzuka N, Nakamura S, Inuzuka M, Tomoeda M. Specific action of sodium dodecyl sulfate on the sex factor of Escherichia coli K-12 Hfr strains. J Bacteriol 1969; 100: 827-835.
- Aja AM, Gasca AG, Grobois AA, Mejia CB, Roque A, Gil BG. Plasmid profiling and antibiotic resistance of Vibrio strains isolated from cultured penaeid shrimp. FEMS Microbiol Lett 2002; 213: 7-12.
- Schaufler K, Wieler LH, Semmler T, Ewers C, Guenther S. ESBL-plasmids carrying toxin-antitoxin systems can be "cured" of wild-type Escherichia coli using a heat technique. Gut Pathog 2013; 5: 34.
- Dale JW, Park SF: Molecular genetics of bacteria (5th Edn). John Wiley & Sons, New York, Oxford, 2010.
- Nasreen J, Sudhir U. Meshram, Archana K. Plasmid profile analysis of multidrug resistant E. coli isolated from UTI patients of Nagpur City, India. Rom Biotechnol Lett 2009; 14: 4635-4640.
- Suhani S, Purkaystha A, Begum MK, Islam MJ, Azad AK. Plasmids for amoxicillin and ciprofloxacin resistance in Escherichia coli isolate causing urinary tract infection. Clin Microbiol 2017; 6: 284.
- Reboucas RH, Viana de Sousa O, Sousa LA, Roger FV, Carvalho PB, Fernandes RHV. Antimicrobial resistance profile of Vibrio species isolated from marine shrimp farming environments (Litopenaeus vannamei) at Cear’a, Brazil. Environ Res 2011; 111: 21-24.
- Albuquerque Costa R, Araújo RL, Souza OV, Vieira RH. Antibiotic-resistant vibrios in farmed shrimp. Biomed Res Int 2015; 2015: 505914.
- Liu X, Wang D, Wang H, Feng E, Zhu L. Curing of plasmid pXO1 from Bacillus anthracis using plasmid incompatibility. PLoS ONE 2012; 7:e29875.
- Carvalho AC, Barbosa AV, Arais LR, Ribeiro PF, Carneiro VC, Cerqueira AM. Resistance patterns, ESBL genes, and genetic relatedness of Escherichia coli from dogs and owners. Braz J Microbiol 2016; 47: 150-158.
- Stenske KA, Bemis DA, Gillespie BE. Comparison of clonal relatedness and antimicrobial susceptibility of fecal Escherichia coli from healthy dogs and their owners. Am J Vet Res 2009; 70:1108-1116.
- Casellas JM. Antibacterial drug resistance in Latin America: consequences for infectious disease control. Rev Panam Salud Publica 2011; 30: 519-528.