- Biomedical Research (2014) Volume 25, Issue 4
Analysis of Cytokine Gene Polymorphism Allelic Variation in the Turkish Population with Inflammatory Bowel Disease.
Ilhami Gok1*, Fahri Uçar2, Orhan Ozgur3 and Hasan Efe41Department of Bioengineering, Faculty of Engineering & Architecture, Kafkas University, 36100, Kars, Turkey
2Departments of Medical Biology & Genetics, School of Medicine, Akdeniz University, 07200, Antalya, Turkey
3Karadeniz Technical University, School of Medicine, Department of Internal Medicine, Divisions of Gastroenterology, 6100, Trabzon, Turkey
4Department of Biochemistry, School of Medicine, Recep Tayyip Erdoğan University, 53300, Rize, Turkey
- *Corresponding Author:
- İlhami GÖK
Department of Bioengineering
Faculty of Engineering & Architecture
Kafkas University, 36100, Kars, Turkey
Accepted date: June 11 2014
Abstract
Inflammatory bowel diseases (IBD), Crohn’s Disease (CD) and Ulcerative Colitis (UC) are complex diseases showing genetic heterogeneity that arise with the interaction of the genetic and environmental agents. The prevalence of the cytokine gene polymorphism has different distributions in the various races of the world. In our study, 22 polymorphic regions of 13 different cytokine genes were evaluated in IBD patients in the Black Sea Region of Turkey. Sixty-nine IBD patients (18 with Crohn’s disease and 51 with ulcerative colitis) and 100 healthy individuals as controls who lived in the Black Sea Region and whose clinical diagnoses were realized in Research Hospitals, were selected. DNA was isolated from the blood of the selected individuals and the analyses of the cytokine gene polymorphisms were determined with the method of PCR-SSP (sequence specific primer). When the patients were compared with the control group in terms of cytokine genes, significant changes were observed in the allelic frequencies of the diseased group compared to the healthy individuals thus implying a role of allelic frequencies in Crohn’s disease and ulcerative colitis. We conclude that, the allelic changes arising at the genomic level in the cytokine gene polymorphisms in the Turkish population could make contributions to the clinic diagnosis of patients with IBD.
Keywords
Inflammatory bowel disease , cytokine gene polymorphism, PCR-SSP, Black Sea Region, Turkish population
Introduction
Inflammatory bowel diseases (IBD) are in the group of chronic inflammatory diseases having the indications such as abdominal cramp, rectal bleeding and diarrhea [1]. Crohn’s disease (CD) and ulcerative colitis (UC) are two main forms of the inflammatory bowel disease having multi-factorial inheritance characteristics arising with the interaction of genetic and environmental factors [2]. There is a continuing trend towards an increasing incidence and prevalence of IBD across Asia (particularly in East Asia). While this is occurring among developing nations, it is also being seen in Japan, a socioeconomically advanced country [3]. It attracts attention that the incidence and prevalence of IBD have shown significant increase in recent years in Turkey [4]. The distribution of both diseases is different in terms of age, geography, race, ethnic, and social segments [5]. In clinical cases, environmental factors, infecting agents, increase of permeability in the small bowel, autoimmune responses, and auto antibodies were also involved in these illnesses in addition to the genetic factors [6]. It is thought that the cytokines, which mediate inflammation in IBDs have many functions in the immune system. Also, it was determined by the researchers that the cytokines could be important in the pathogenesis of IBD [7-9]. Studies in this field have shown that with the negative changes of the environmental conditions cytokine gene fluctuations increase and this triggers IBD [10]. In IBD patients, the relationship between the candidate gene polymorphism and sensitiveness to this illness was proposed to differ among different population. In this study, we investigated the cytokine gene polymorphisms in the Black Sea Regional population and to determine the cytokine gene polymorphism changes in IBD at the allelic level.
Materials and Methods
Patients and genomic DNA extraction
This research was conducted among the Black Sea Region population, which geographically corresponds to Northern Turkey. Sixty-nine cases for this study were selected from the patients applying to the Trabzon Farabi Hospital Gastroenterology and Kocaeli Medicine Faculty Gastroenterology polyclinics with inflammatory bowel diagnosis and 100 healthy individuals as control group. IBD was determined using the physical examination, radiological, endoscopic, and pathological test criteria according to the complaints of the patients. From each of the 69 patient and 100 healthy individuals included in the study, 8 ml blood was taken into EDTA tubes. From the peripheral blood taken, DNA isolation was conducted with the method of Salting – Out protocol [11]. The optical density of the concentration of DNA obtained in this stage was read in the Nanodrop spectrophotometer (Thermo) in 260 nm wave length [12]. The isolated DNA samples were kept at -80°C. The genomic analyses were completed in School of Medicine, Hematology Department tissue typing Laboratory.
PCR-SSP and electrophoresis
The Protrans cytokine Cycler Plate 200096; the Cytokine gene panel, which was obtained from the Protrans firm for the determination of the cytokine gene polymorphism was used to determine 22 polymorphic varieties of 13 different cytokines. It is composed of 3 different sub-components; a) plates of 96 saturated with the oligonucleotide primers, b) R buffer and c) Y buffer. For the PCR-SSP amplification of each individual, the master-mix and DNA mixture having total volume of 521,3 l [Buffer R 138 l, Buffer Y 280 l, Genomic DNA (50-300 ng/l) or 100 l and Taq Polimeraz (5U/l): 3,3 l] was prepared [13].
The prepared master-mix was slightly mixed in the vortex and was distributed to 44 wells in the plate with the final volume being 10 l for each patient. For the following ones given for the cytokine genes, the beginning denaturation was amplified at 94°C for 2 min, denaturation was amplified at 94°C for 10 sec, annealing- extension was amplified at 65°C for 1 min 10 cycles, the final denaturation was amplified at 94°C sec, annealing was amplified at 61°C for 50 sec, the extension was amplified at 72°C for 30 sec, 30 cycles and hold was amplified at 4°C in indefinite PCR cycle programmed. For the implementation of the amplified PCR products in the electrophoresis, 1XTBE buffered %2 agarose gel was prepared. The samples were executed in 1X TBE buffered environment at 75V, 50 A for 35 minutes by being loaded to 44 well gel for each patient (Perrey et al.2002). The results were executed in 1X TBE buffered environment at 75 V , 50 A for 35 minutes by being loaded to 44well gel for each patient. The result were examined in the trans illuminator at UV and the wells having double bands from the samples available in each well were accepted as positive and photographed. The changes in the cytokine gene polymorphisms of the patients were determined as a result of the comparison with the control groups within the information in the chart in the protrans cytokine 2 cycler plate system analysis table [13].
Statistical analysis
The statistical analysis of the cytokine gene polymorphisms was conducted by using the SPSS Windows 16.0.0 (Statistical package for social science). The statistical significance between the groups was calculated by using Pearson’s adjusted Chi- square and Fisher’s exact Chi- square tests. The significance level was selected as p=0.05 in the statistical comparisons throughout the manuscript.
Results
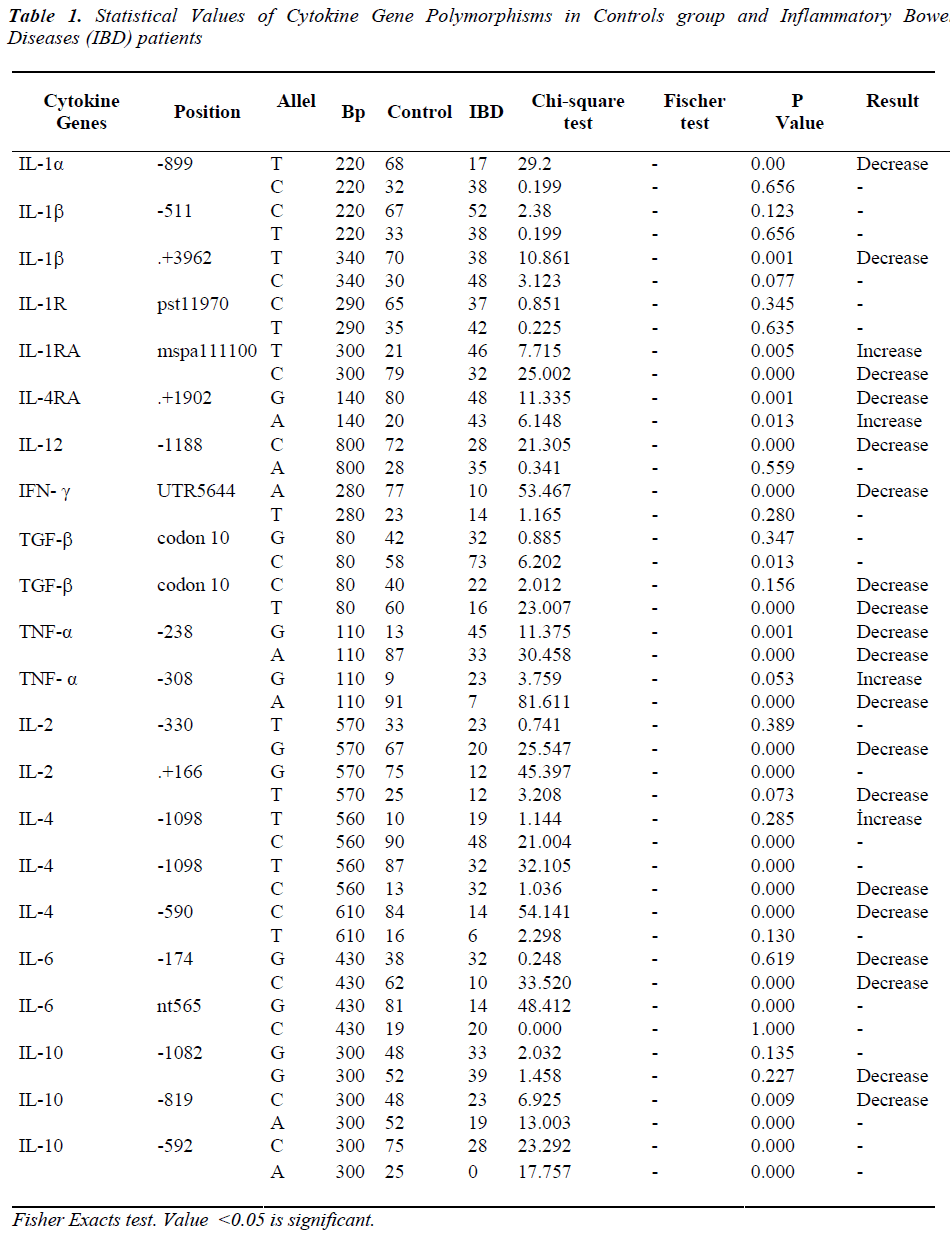
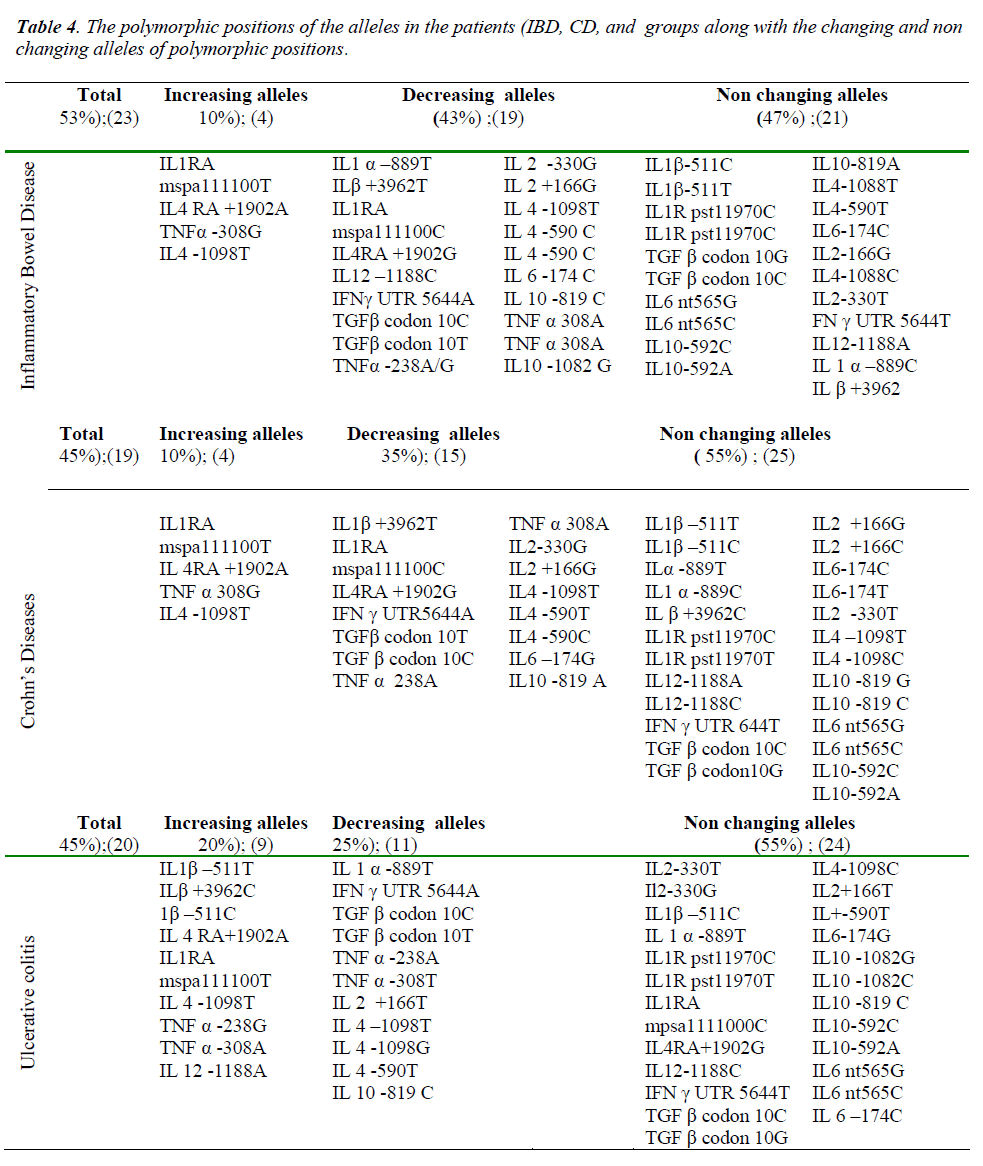
In total, we studied 44 sites of 22 different polymorphic regions of 13 cytokine genes in IBD. The 13 genes are interleukin-1alpha (IL-1α), interleukin-1beta (IL- 1β), interleukin-1reseptor (IL-1R), interleukin-1 antireseptor (IL-1RA),interleukin-4 anti receptor (IL- 4RA), interleukin-2(IL-2), interleukin-4 (IL-4), interleukin- 6 (IL-6), interleukin-12 (IL-12), interferon gamma (IFN-γ), tumor growth factor beta (TGF-β),and tumor necrosis factor alpha (TNF-α) and interleukin-10 (IL-10) . Of the 44 sites examined, a total of 23 (53%) sites in the IBD manifested changes in allele frequencies compared to the control group.
Among those changes, only four (10%) sites had an elevated allele frequency and the other 19 (43%) sites had a decreasing allele frequency compare to the control group allele frequencies (Table 1).
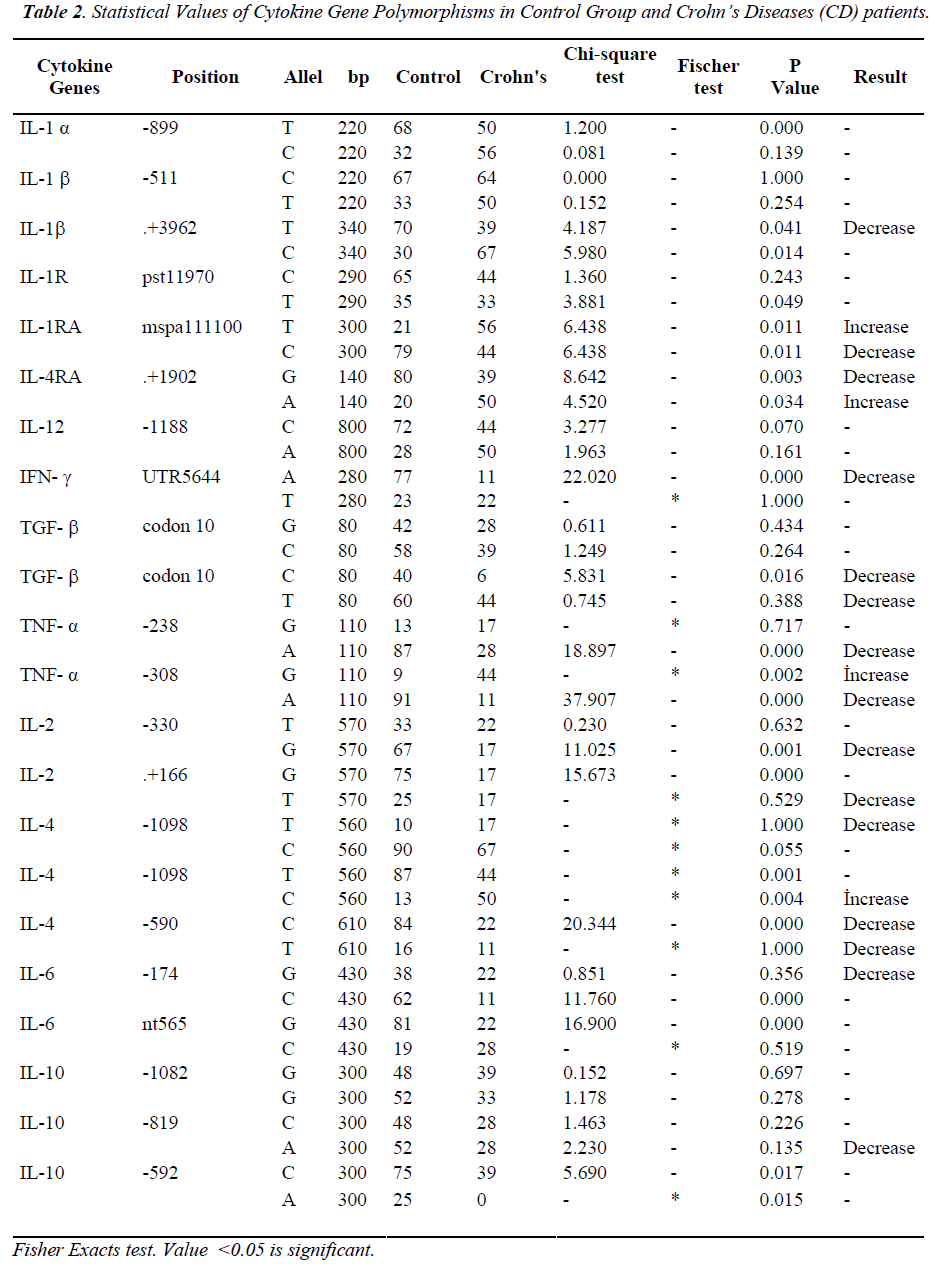
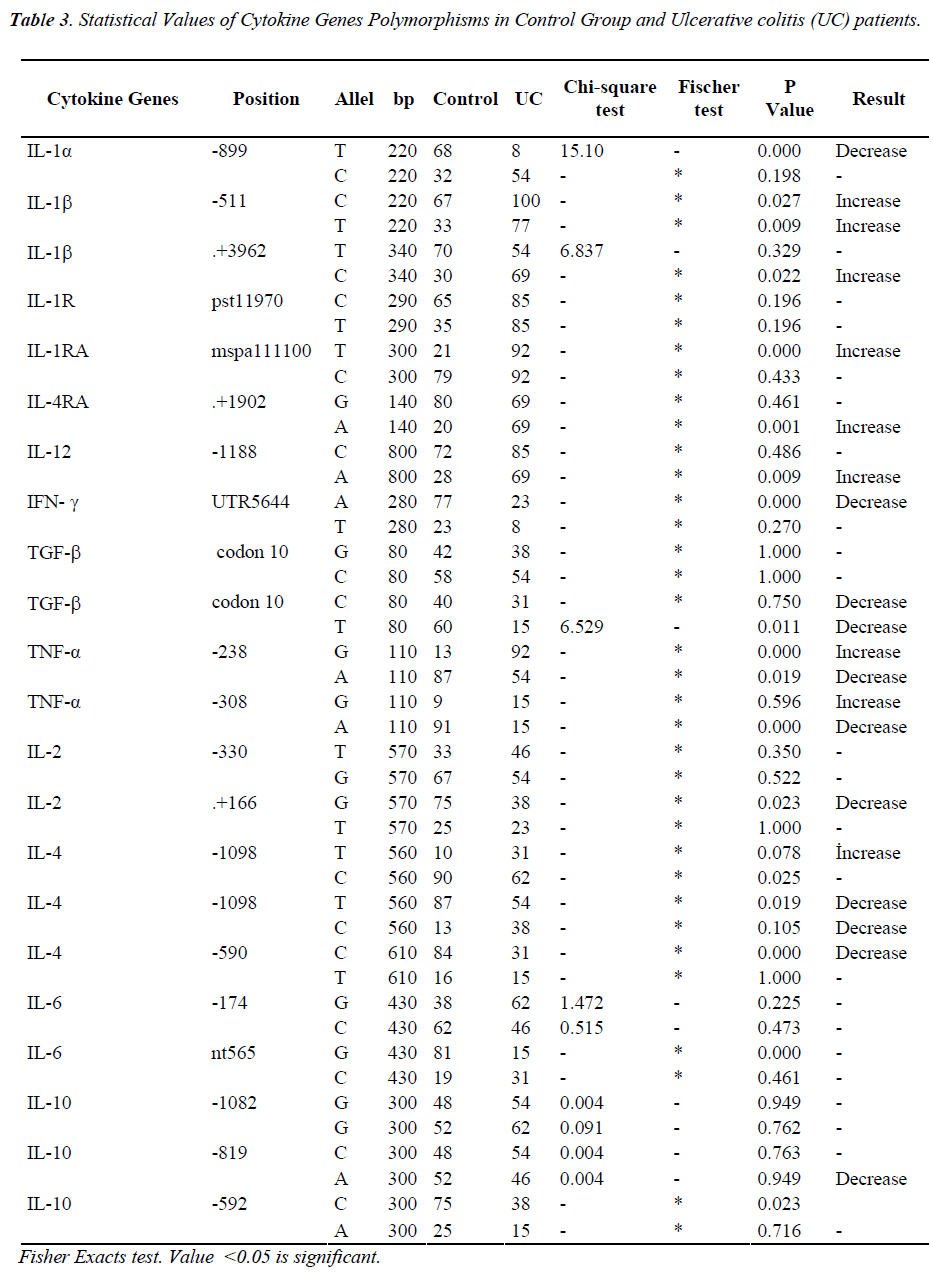
The total number of sites that had changes in allele frequency of the cytokine gene was 19 (about 44%) in terms of Crohn’s disease patients in which 15 (35%) sites showed a decrease and four (10%) sites manifested increase (Table 2). We also observed that, for the ulcerative colitis the total change in cytokine gene polymorphisms was about 20 (45%) sites in which 11 (25%) sites showed decrease in allele frequency compare to the controls and 9 (20%) showed increase (Table 3). Total changes and alleles polymorphic positions were expressed in three groups (IBD, CD and UC) and control group (Table 4).
Discussion
In this study we found that there are certain sites in the cytokine genes that had variegated polymorphic allele frequencies in IBD diseases. We conducted in depth analyses to find out the nature of the changes in the allele frequency of the each site and whether those changes could be used in clinical application of the diagnosis of the disease. Since the allele frequencies are results of accumulation of the mutants in the population, the frequency of alleles can vary in different populations [14-16]. Among the polymorphic sites studied, TNF α -308G site was shown to increase the inflammation and IL -1RA site represses the inflammation. A similar study conducted in the Japan population indicated that the allelic differences of TNF-α and IL- 1 RA genes associated with the inflammation and TNF-α was an important candidate gene in the inflammatory bowel diseases [17]. When the results of our study are compared with the Japanese population, there is a similarity in the allele frequency patterns of TNF-α and IL-1RA genes. Nevertheless, polymorphism pattern of IL-4RA gene obtained in our study shows differences when compared to a number of other studies [18].
To investigate the possible genes associated with Crohn’s disease, wide genome screenings were conducted recently and the effects of the allelic frequency changes in the polymorphic regions were discussed [19,20]. In our study, we have observed that the changes in the allele frequencies in the cytokine gene polymorphisms were less significant in CD and more significant in UC. An existence of a balance between the IL-1α and IL-1β cytokine proinflammators were thus proven. With an increase in the rate of IL-1 (IL-1α and IL-1β), there is inflammation in the mucosa of the patient’s bowel. However, this is not the case in the healthy individuals [21]. Among the US and European population, the irregularity of proinflammatory cytokines (IL-1α, TNF-, IL-12, IL-8 and IL-6) and anti–inflammatory cytokines (IL-4, IL-10, IL- 1RA and IFN-γ) in IBD were reported with an increase in allele frequency [22]. Nonetheless, we conversely observed a decreasing pattern in IBD. We propose that this difference was due to the nature of the population studied. In IBD, the increases in the allele frequency of the TNF- , IL-1, IL-2, IL-6 and IL-8 was with respect to the expression in bowel mucosa [23]. The elevation of allele frequency of the certain sites of TNF- and IL-1, IL-2, IL-6 and IL-4 significantly increased in the bowel mucosa in UC and CD in the Chinese population [24,25]. In our research, in the cytokine gene polymorphisms of UC patient, there were increases in IL-1 β –511C, IL-1 β – 511T, IL-1 β +3962C, and decreases in IL -1 α -889C, IL- 1 β +3962C, IL -1RA mspa 111100T, and no changes in the others. In UC, the repetitions of the IL-1 RA gene polymorphisms have significantly increased. This increase was observed in Ashkenazi Jews in the state of Los Angeles [19]. In IBD, it was shown that there were decreases in the allele frequencies of the same gene and allelic frequencies of IL-1RA were shown to increase between UC and CD patients [26]. The same pattern was observed in the patients with IBD [27]. Some studies showed that some cytokines (IL-1 and IL-1RA) play roles in the formation of the disease and in the development of the inflammation in IBD [27]. Our findings here are in parallel with the studies conducted in various populations in terms of IL-1RA and TNF-α. It is reported that frequency of some alleles in the IL-1RA has significantly increased in the patients with CD and UC and the elevation leaded the increase in the formations of the lesions [28].
The effects of polymorphisms in IL-1 and IL-1 RA genes in the Europe population is associated with UC, CD, and healthy control groups and the results revealed that there is an association for one single positive allele of IL-1 gene for CD and two positive alleles for IL-1RA [29]. Scientists showed that there was an important decrease in the rate of IL-1/IL1-RA in the IL-1 and IL-1RA measurements that they have realized in UC, CD and control groups [30]. However, we our results differ with previous studies in terms of allele frequencies of the IL- 4RA. We found that the allelic frequencies of IL-4RA as between 60-69% in UC and as 39-50% in the CD patients in the regional Turkish Community [4]. In the recent research, the hypothesis that the IBD can be treated via cytokines and for this reason, the activity, excretion and synthesis of the cytokines and cytokine signal pathways in the target cells in the bowel mucosa immune system can be inhibited [31,32]. In our research, however, a decreasing, not an increasing pattern in the IL-2, IL-4, IL-6 polymorphisms in IBD, CD, and UC was observed.
In conclusion, we observed the cytokine genes that vary in the direction of increasing frequencies of allelic genes (eg), while IL -1RA , IL - 4RA , TNF - α and IL - 4 is similar in all the three disease groups (IBD , CD and UC) . However, UC allelic frequency of IL- 1β exhibit significant increase that maybe diverge from CD and IBD phenotypically. Pro-inflammatory cytokine, IL- 1β gene may have induced the inflammation in the UC. Moreover, this result may then be potentially used at the UC clinic to differentiate patients with UC from IBD and CD. In general, pro-inflammatory cytokines (e.g. TNF - α and IL - 4) and anti -inflammatory cytokines (IL -1RA , IL - 4RA) regulate themselves during an inflammatory reaction either by increasing or decreasing its response, respectively. Analyses of the cytokine genes in this study showed that the change in the allelic frequencies in the lower direction of the IL - 1α and TGF - β , TNF - α , IL - 2 , IL - 4 , IL - 10 and IFN - Ɣ was relatively high in all disease groups (IBD , CD and UC). High rate of reduction in the allelic frequencies of cytokine genes in these groups showes that anti-inflammatory cytokines and inflammatory diseases play an important role in the diagnosis . Overall, to the best of our knowledge, We think that the changes in the direction of increasing and decreasing in the cytokine gene polymorphisms can be a factor contributing to the formation of IBD. The changes could be used in the clinical diagnosis of the patients. In addition to the formation of IBD, determination of cytokine gene polymorphism in IBD could ultimately help for clinical diagnosis of cancer, autoimmune disease, and other multifactorial genetic disorders resulting from inflammation. We have a positive approach to the idea that in addition to the effects of the candidate genes in the formation of the above mentioned diseases, different environmental conditions, genotypic differences in the regional populations, nutritional habits, and geographical factors could play a role.
Acknowledgments
We are grateful to the Karadeniz Technical University Scientific Research Project unit (Trabzon, Turkey.Grant : No: 22.114.001.11) for financially supporting this study.
References
- Franke A, Fischer A, Nothnagel M, et al. Genome-wide association analysis in sarcoidosis and Crohn's disease unravels a common susceptibility locus on 10p12. Gastroenterology 2008; 135: 1207-1315.
- Yamamoto-Furusho JK, Santiago-Hernández JJ, Perez-Hernandez N, et al. Interleukin 1β (IL-1B) and IL-1 antagonist receptor (IL-1RN) gene polymorphisms are associated with the genetic susceptibility and steroid dependence in patients with ulcerative colitis. J Clin Gastroenterol 2011; 45: 531-535.
- Bernstein CN. Inflammatory Bowel Diseases: Global Perspective.World Gasroenteroloji Organization Guidline Publishing 2009; 1: 3-5.
- ÇelikY, Dağlı Ü, Yalın Kılıç M, et al. Cytokine gene polymorphism in Turkish patients with inflammatory bowel disease. Scand J Gastroenterol 2006 ; 41: 559-565.
- Doecke JD, Simms LA, Zhao ZZ, et al. Genetic susceptibility in IBD: overlap between ulcerative colitis and Crohn's disease. Inflamm Bowel Dis 2013; 19: 240-245
- Yamamoto-Furusho, JK, De-León-Rendón, JL, de la Torre MG, et al. Genetic polymorphisms of interleukin 20 (IL-20) in patients with ulcerative colitis.Immunol Lett 2013; 149: 50-53.
- Arisawa T, Tahara T, Shibata T, et al. The influence of polymorphism of interleukin-17 and interleukin-17 F genes on the susceptibility to ulcerative colitis. J Clin Immunol 2008; 28: 44-49.
- Lu C, Waugh A, Bailey RJ, et al. Crohn's disease genotypes of patients in remission vs relapses after infliximab iscontinuation. World J Gastroenterol 2012; 28: 18: 5058-5064.
- Ferguson LR, Huebner C, Petermann I, et al. Single nucleotide polymorphism in the tumor necrosis factoralpha gene affects inflammatory bowel diseases risk. World J Gastroenterol 2008; 14: 4652-4961.
- Schmidt C, Giese T, Goebel R, et al. Interleukin-18 is increased only in a minority of patients with active Crohn’s disease. Int J Colorectal Dis 2007; 22: 1013-1020.
- Rapley R, Walker JM. The Nucleic Acid Protocol Handbook. Humana Press, Ottowa, New Jersey 2008;pp 9-27
- Zhu H, Lei X, Liu Q, et al.Interleukin-10-1082A/G polymorphism and inflammatory bowel disease susceptibility: a metaanalysis based on 17,585 subjects. Cytokine 2013; 1: 146-1453.
- Perrey C, Pravica V, Sinnott PJ, et al. Genotyping for polymorphism in interferon gamma,interleukin-10,TGF beta-1 and TNF-alfa genes: a technical report, Transplant İmmunology 2003: 6: 193-197.
- Bashashati M, Rezaei N, Bashashati H, et al.Cytokine gene polymorphisms are associated with irritable bowel syndrome: a systematic review and meta-analysis.Neurogastroenterol Motil 2012; 12: 1102-1106
- Jiang Y, Lin XQ, Cao SG, et al. Correlations of genetic polymorphisms of tumor necrosis factorrelated apoptosis-inducing ligand gene and its plasma phenotype with ulcerative colitis. Chinese 2012; 18: 1244-1248.
- Walczak A, Przybylowska K, Dziki L, et al. The lL8 gene polymorphisms in inflammatory bowel diseas e and colorectal cancer.DNA Cell Biol 2012; 8: 1431-1438.
- Hollegaard MV, Bidwell J L. Cytokine gene polymorphism in human disease: on-line databases, Supplement 3. Genes Immun 2006; 7(4): 269-276.
- Lu Z, Chen L, Li H, et al. Effect of the polymorphism of tumor necrosis factor-alpha-308 G/A gene promoter on the susceptibility to ulcerative colitis: a metaanalysis. Digestion 2008; 78: 44-51.
- Neuman MG, Nanau RM. Single-nucleotid polymorphisms in IBD. Transl Res 2012; 160: 45-64.
- Moran CJ, Walters TD, Guo CH, et al. IL-10 polymorphisms are associated with very-early-onset ulcerative colitis.Inflamm Bowel Dis 2013; 19: 115-123
- Glas J, Seiderer J, Wagner J, et al. Analysis of IL12B gene variants in inflammatory bowel disease. PLoS One 2012; 7: e34349.
- Liberek A, Jakóbkiewicz-Banecka J, Kloska A, et al. Clinical parameters of inflammatory bowel disease in children do not correlate with four common polymorphisms of the transforming growth factor β1 gene.Acta Biochim Pol 2011; 58: 641-644.
- Marrakchi R, Moussa A, Ouerhani S, et al.Interleukin 10 promoter region polymorphisms in inflammatory bowel disease in Tunisian population. Inflamm Res 2009; 58: 155-160.
- Li K, Yao S, Liu S, et al. Genetic polymorphisms of interleukin 8 and risk of ulcerative colitis in the Chinese population. Clin Chim Acta 2009; 405: 30-34.
- Yu P, Shen F, Zhang X, et al. Association of single nucleotide polymorphisms of IL23R and IL17 with ulcerative colitis risk in a Chinese Han population.PLoS One 2012;7: 44380.
- Santana G, Bendicho MT, Santana TC, et al.The TNF-α -308 polymorphism may affect the severity of Crohn's disease.Clinics 2011; 66: 1373-1378.
- Corleto VD, Pagnini C, Margagnoni G, et al. IL-1beta-511 and IL-1RN*2 polymorphisms in inflammatory bowel disease: An Italian population study and metaanalysis of European studies. Dig Liver Dis 2010 :42: 179-184.
- Amre DK, Mack DR, Morgan K, et al. Interleukin 10 (IL-10) gene variants and susceptibility for paediatric onset Crohn's disease. Aliment Pharmacol Ther 2009; 29: 1025-1031.
- Haukim N, Bidwell JL, Smith AJP, et al. Cytokine gene polymorphism in human disease: On-line databases. Genes İmmun 2002; 3: 313-330.
- Ben Aleya W, Sfar I, Habibi I et al. Interleukin-18 gene polymorphisms in tunisian patients with inflammatory bowel disease. Digestion 2011; 83: 269-274
- Hong J, Leung E, Fraser AG, et al. IL-4, IL-10, IL-16 and TNF polymorphisms in New Zealand Caucasian Crohn's disease patients. Int J Colorectal Dis 2008; 23: 335-337.
- Liang WD, Li JS, Li KS, et al. IL-8 gene polymorphisms with inflammatory bowel disease in Chinese patients. Chinese 2001; 26: 1825-1829.