- Biomedical Research (2013) Volume 24, Issue 3
Analysis of bacterial diversity in healing and non-healing wounds among Malaysian subjects by phenotypic identification and 16S rDNA sequencing.
Shinyee Wong1, Rishya Manikam2, Sekaran Muniandy1,*1Department of Molecular Medicine
2Department of Trauma and Emergency, Faculty of Medicine, University of Malaya, 50603 Kuala Lumpur, Malaysia
- *Corresponding Author:
- Sekaran Muniandy
Department of Molecular Medicine
Faculty of Medicine, University of Malaya
50603 Kuala Lumpur Malaysia
Accepted May 03 2013
Abstract
Wound healing is a complex sequence of events consisting of regeneration and repair. It has been proposed that bacteria present in wounds play a major role in delayed wound healing. The main objectives of this study are to identify the bacterial species present in wounds and to evaluate and compare the results obtained using conventional biochemical analysis and 16S rDNA sequencing. Wound swabs from 15 healing and 15 non-healing wounds were collected from the wound clinic in University Malaya Medical Centre, Malaysia. The wound microbiotas obtained by bacterial culture methods followed by biochemical tests and 16S rDNA sequencing were compared. Culture analysis and 16S rDNA sequencing both revealed that the most prevalent bacteria in wounds are Staphylococcus spp. and Pseudomons spp. The prevalence of Staphylococcus aureus was significantly higher in non-healing wounds than healing wounds (p=0.003). API® biochemical tests successfully identified all 54 isolates at species level. However, by 16S rDNA sequencing only 19 (35.2%) isolates were identified at species level and 35 (64.8%) were identified at the genus or family level. 16S rDNA sequencing however provided a better view on the phylogenetic relationships of bacterial species present in wounds.
Keywords
Bacterial identification, Biochemical tests, 16S rDNA sequencing
Introduction
Intact skin plays a major role in controlling microbiota on the skin surface and prevents underlying tissue from being colonised and invaded by potential pathogens. External damage to the intact skin through wounding provides a favourable environment for microbial colonisation and proliferation [1]. Wound healing is a complex process characterised by three consecutive and overlapping stages: inflammation, proliferation and remodelling [2]. Wound healing begins with platelet aggregation that promotes haemostasis. This is followed by an inflammatory cell cascade involving neutrophils, macrophages and lymphocytes within the tissue and lastly closure of wound via proliferation of fibroblasts and remodelling of extracellular matrix by crosslinking of collagen and scar maturation [3,4]. Proper circulation, nutrition, immune state and avoidance of negative mechanical forces are necessary for normal wound healing. Alteration in any of these will lead to a delay in wound healing [5].
There are two types of wounds: acute and chronic. Acute wounds are caused by external trauma such as surgical wounds, bites, burns, minor cuts, abrasions, crushing and gun shoot injuries [1]. Acute wounds usually heal in a very orderly and efficient manner within a predictable time frame that leads to recovery of anatomical and functional integrity [3,6]. Chronic wounds, however do not heal in a predictable period and are biologically defined by prolonged inflammation, defective re-epithelisation and impaired matrix remodelling [7]. These wounds are mostly caused by endogenous mechanisms associated with predisposing conditions such as venous disease, diabetes mellitus, central nervous system compromise, trauma, inflammatory illnesses, metabolic abnormalities, coagulopathies, immunosuppression, obesity, smoking and malnutrition [8]. The presence of non-replicating microorganisms in a wound may be considered as contamination [9]. Contaminating microorganisms arise from three main sources: the external environment (air or those introduced by traumatic injury), the surrounding skin (normal skin microfloras including Staphylococcus epidermidis, Micrococci) and endogenous sources including the gastrointestinal, vaginal and oral tracts [1]. Contamination might proceed to colonisation then local infection and ultimately cause systemic disease such as cellulitis, septicaemia or endocarditis [7,10]. Complications of infection maybe caused directly (bacteria patho genicity) or indirectly (mediated by the immune response in an attempt to eradicate the invading microorganisms) [4].
In the last decade, studies of universal DNA sequences, especially the small ribosomal subunit 16S rDNA of bacterial genes played an important role in the accurate identification of bacterial species and the discovery of novel bacteria in clinical microbiology [11,12]. The 16S sequence consists of highly conserved nucleotide sequences which are present in all bacterial species and variable regions that are genus- or species specific. DNA from almost any bacterial species can be identified by using universal PCR primers which target the conserved regions. Bacterial species can then be revealed by comparing the sequences of PCR products with the known sequences in GenBank or other databases [13].
In this study, we prospectively enrolled 15 patients with healing wounds and 15 patients with non-healing wounds from the Wound Clinic, UMMC. The prevalence of bacterial species in the wounds was determined. Two sets of 16S rDNA primers, PL06 and DG74 were evaluated for their ability to identify bacteria from wound. Identity was compared with that obtained by biochemical tests.
Materials and Methods
Sample Collection and Isolation of Bacteria
Wound swabs of 15 healing wounds and 15 non-healing wounds were collected from subjects attending the Wound Clinic in University Malaya Medical Centre through a study approved by the Ethic Committee of UMMC (Ethics Number: 962.13). Using sterile technique, a cotton swab was rotated over the wound for 5 seconds and the tip of the swab was then broken into a sterile transport tube containing 3ml of Phosphate Buffered Saline. 100μl of each sample was spread on Mac- Conkey agar and Phenylethyl Alcohol Sheep Blood Agar. The plates were then incubated aerobically and anaerobically at 37ºC for 4 days. Pure cultures were obtained after several plates were streaked with single colony.
Biochemical Methods
Pure isolates were phenotypically identified through Gram Stain and biochemical tests. API®Coryne was used for Gram-positive bacilli, API®Strep was used for catalase negative Gram-positive cocci, API®Staph was used for catalase positive Gram-positive cocci, API®20E was used for oxidase negative Gram-negative bacteria and API®20NE was used for oxidase positive Gramnegative bacteria. Identification was carried out according to the manufacturer’s instructions. After 24-48 hours incubation, the strip was read according to the reading table and the interpretation of the data was done by using the bacterial identification software (APIwebTM).
16S rDNA Sequencing
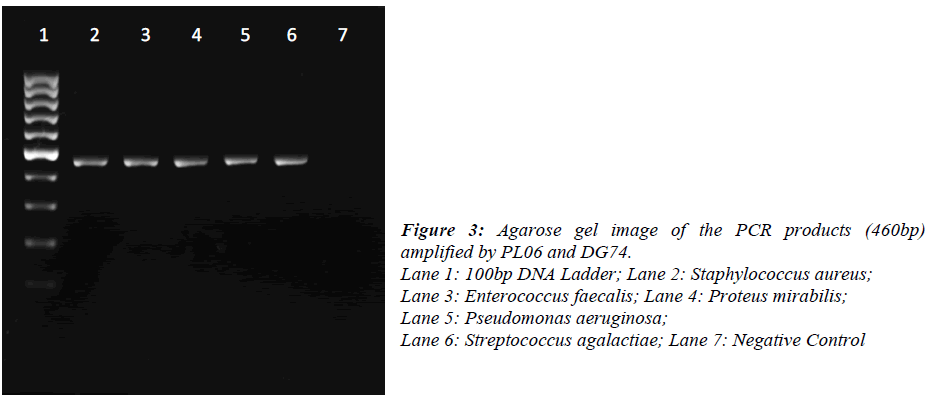
DNA was extracted from pure bacterial colonies using igenomic DNA extraction kit (iNTRON Biotechnology, Korea). A single colony was picked up from the agar plate and re-suspended into 1.5ml Phosphate Buffered Saline. Bacteria were pelleted by centrifugation at 11337xg for 1min and suspended in 100μl of Buffer MP and 3μl of lysozyme solution. The extraction was performed according to the manufacturer’s instruction. Universal primers for 16S rDNA (PL06: 5’- GGT TAA GTC CCG CAA CGA GCG C -3’ and DG74: 5’- AGG AGG TGA TCC AAC CGC A -3’) were used for PCR amplification. The reaction was carried out in 25μl containing 12.5μl of 2X PCR master mix (Promega, USA), 2.5μl of each primer (10μM), DNA template (1ng) and nuclease free water. PCR conditions were: initial denaturation at 95ºC for 5 min, 30 cycles of denaturation at 95ºC for 1 min, annealing at 55ºC for 1 min, extension at 72ºC for 1 min and final extension at 72ºC for 10 min. Amplification was performed on a Eppendorf Mastercycler Personal (Eppendorf, Germany).16S rDNA sequencing was performed on a ABI 3730XL (First Base Sdn Bhd, Malaysia). The obtained sequences were analysed by using Megablast®. The resulting 16S rDNA sequences of approximately 460 bases were compared with the sequences in National Centre for Biotechnology Information (NCBI) using the BLAST® program. Bacterial were identified on the basis of at least 98% similarity to 16S rDNA sequences in the database
Results
Biochemical Tests and Bacterial Identification
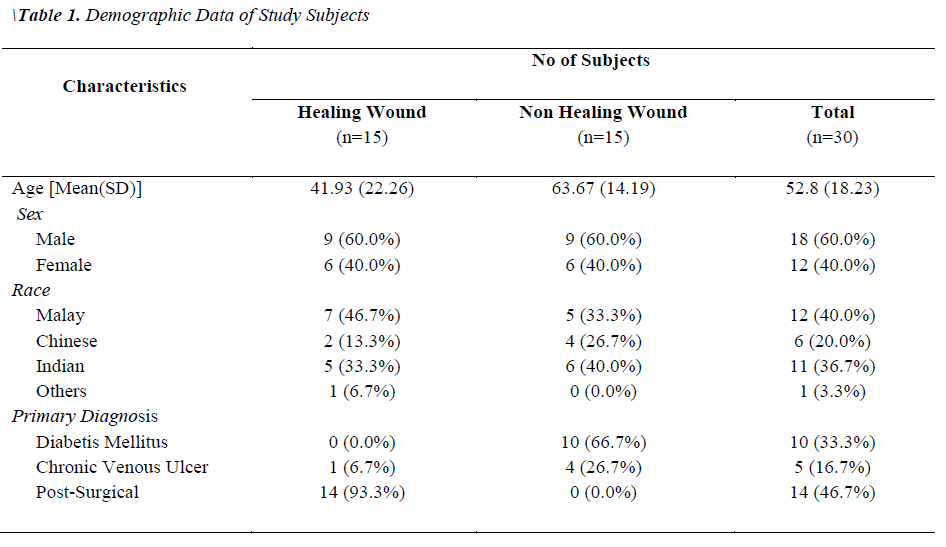
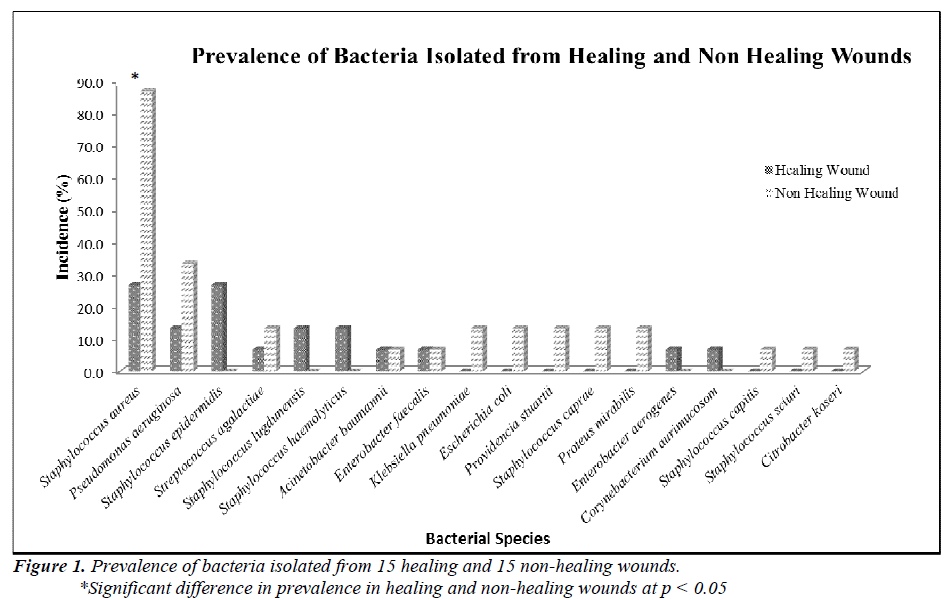
Two groups of patients were recruited in this study. Group I consisted of patients with healing wounds, while group II consisted of patients with non-healing wounds. The demographic and clinical characteristics of the patients are listed in Table 1. The mean age of the healers (41.93±22.26) was significantly lower (p-value 0.0035) than that of non-healers (63.67±14.19). All non-healing wounds were tested positive for at least one organism by biochemical test, while only 60% of the healing wounds showed the presence of bacterial species. A total of 54 clinically relevant bacterial isolates were identified and the results are presented in Figure 1. Coagulase-positive Staphylococcus aureus (56.7%) and Pseudomonas aeruginosa (23.3%) were the most prevalent wound bacteria. The prevalence of Staphylococcus aureus was significantly higher in non-healing wounds than in healing wounds (p=0.003).
Comparison of Bacterial Identification by Biochemical Analysis with 16S rDNA Sequencing
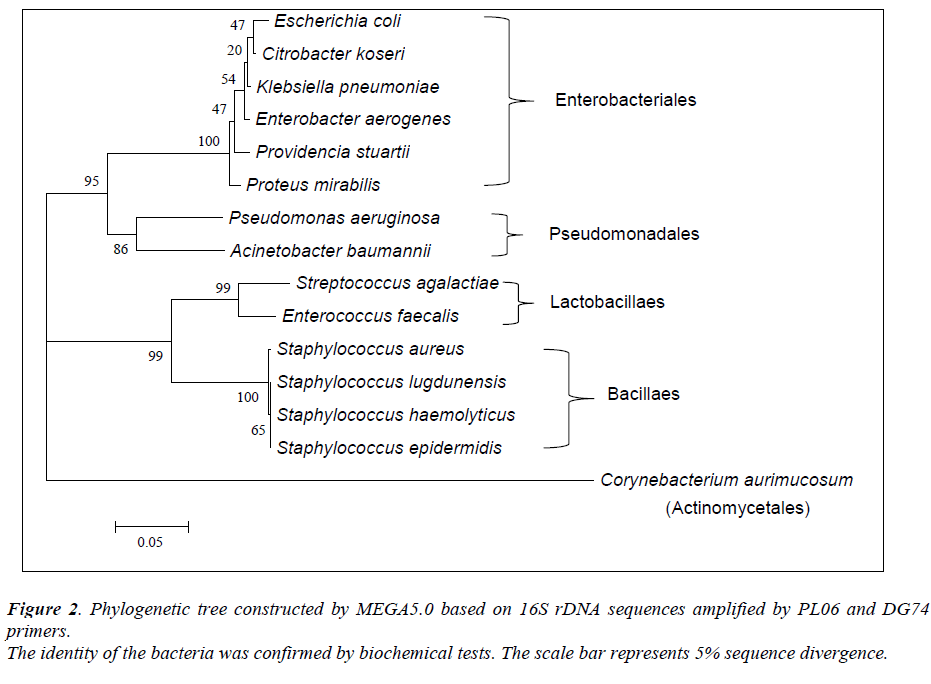
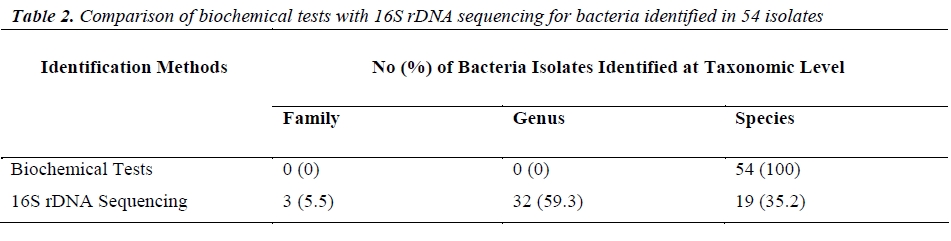
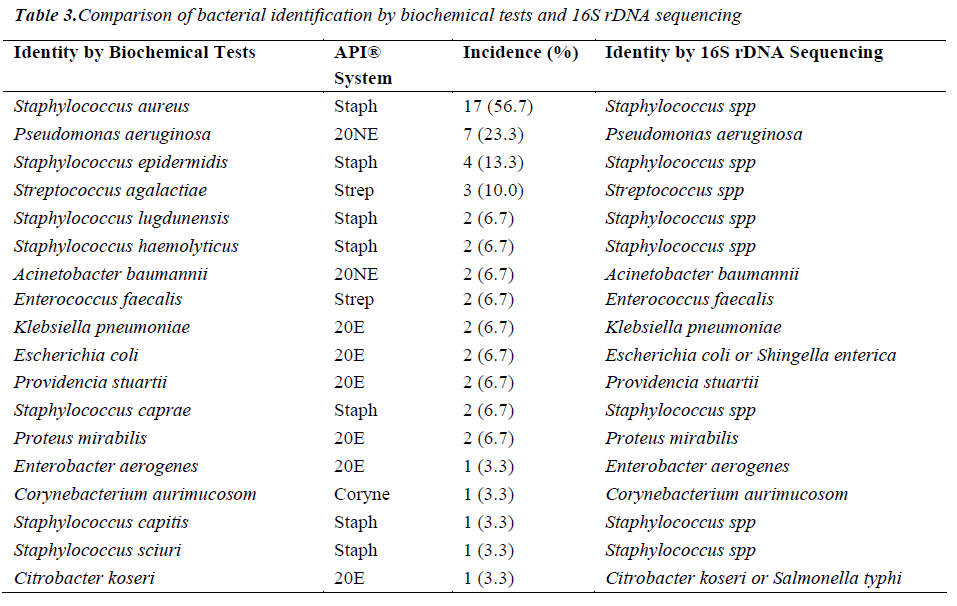
Of the 54 isolates, 29 (53.7%) were identified by API® Staph system, 10 (18.5%) were identified by API®20E system, 9 (16.6%) were identified by API®20NE system, 5 (9.3%) were identified by API®Strep system, and 1 (1.9%) was identified by API®Coryne system. The results obtained were scored as excellent, very good, good and acceptable according to the manufacturer’s guide. Primers DG74 and PL06 located at the highly conserved 16S rDNA region successfully amplified target DNA for all 54 colonies isolated with PCR products of approximately 460 base pairs (Figure 2). While 54 isolates were identified at species level by the API® system, only 19 (35.2%) isolates were identified by 16S rDNA sequencing to the species level, 32 (59.3%) were identified at genus level and 3 (5.5%) were identified at family level (Table 3). For instance, sequence comparison of an isolate with sequences in the NCBI database showing 100.0% concordance to both Staphylococcus aureus and Staphylococcus epidermidis. Hence this isolate can only be identified up to its genus level, Staphylococcus spp. Analysis of the phylogenetic tree revealed that these two organisms are closely joined (Figure 3). For 35 isolates identified to the family and genus level by 16S sequencing, API® assigned the isolates to the same genus and further identified to the species level. The identification of 19 isolates to species level by 16S rDNA sequencing was identical to that identified by the biochemical results
Discussion
Our findings show that the mean age of the non-healers was significantly higher than the healers and this finding is in agreement with the previous findings [14,15]. A study by Harker [16] showed that systemic diseases such as diabetes mellitus, chronic venous ulcers and arterial ulcers which may delay wound healing are more common in elderly. Moreover, factors such as excessive inflammatory response, matrix degradation, impaired vascularization, change in energy metabolism and decreased granulation may also affect wound healing in older subjects [15,17].
Previous studies have shown that Staphylococcus aureus and Pseudomonas aeruginosa are the predominant microorganisms isolated from chronic wounds [4,7,10]. This is in agreement with results from this study in which the prevalence of Staphylococcus aureus was found significantly higher in non-healing wounds than in healing wounds. Athanasopoulos et al. [18] and Edwards et al. [19] postulated that the extracellular adherence protein (Eap) of Staphylococcus aureus played a pivotal role in impaired wound healing by impeding the inflammatory state and inhibiting angiogenesis in the proliferative state. Inflammation is an important part in wound healing and is responsible for eliminating potential pathogens [4]. However, presence of bacterial components in chronic wounds may stimulate excessive inflammatory response and chronic wounds will not heal until the excessive inflammation is reduced [3].
For decades, biochemical methods have been used as the ‘gold standard’ in identification of bacteria. To find a effective and efficient method in identification of wound bacteria, we compared and evaluated both biochemical methods and 16S rDNA sequencing. Our findings revealed that all 54 isolates were identified by the API® systems to the species level. However, only 19 isolates were identified to species level by 16S rDNA sequencing. Previous studies had reported that 16S rDNA sequencing provides genus identification in >90% of the cases and species identification in only 65-83% cases [12,20]. Indeed, the 16S primers (PL06 and DG74) that were used in this study have poor discriminative power among closely related species. For instance, Staphylococcus aureus and Staphylococcus epidermidis showed only <0.5% divergence (>99.5% similarity). Furthermore, construction of a phylogenetic tree revealed little sequence divergence between 16S rDNA sequences in them. Identification of bacteria by 16S rDNA sequencing depends on significant interspecies differences and small intraspecies differences in the 16S regions [12]. Hence, one of the limitations of 16S rDNA sequencing is inability to identify bacterial species particularly when the sequences have very high similarity score to the next closest match [21]. In conclusion, we confirmed that Staphylococcus aureus is the most prevalent bacteria in non-healing wounds of older patients. 16S rDNA sequencing provides us a better view in bacterial phylogeny and taxonomy studies even though there are some limitations in this method. Additional primers or probes are necessary to target the variable region in the 16S sequence for distinctive bacterial identification.
Acknowledgements
We gratefully acknowledge funding for the research described in this study from University Malaya IPPP grant (PG056-2012B). We thank the nurses of the UMMC Wound Clinic for their knowledge and helpfulness.
References
- Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev 2001;14(2): 244-69.
- Rodero MP, Khosrotehrani K. Skin wound healing modulation by macrophages. Int J Clin Exp Pathol 2010; 3(7): 643-653.
- Diegelmann RF, Evans MC. Wound healing: an overview of acute, fibrotic and delayed healing. Front Biosci 2004; 9: 283-289.
- Jones SG, Edwards R, Thomas DW. Inflammation and wound healing: the role of bacteria in the immuno- regulation of wound healing. Int J Low Extrem Wounds 2004; 3(4): 201-208.
- Fonder MA, Lazarus GS, Cowan DA, Aronson-Cook B, Kohli AR, Mamelak AJ. Treating the chronic wound: A practical approach to the care of nonhealing wounds and wound care dressings. J Am Acad Dermatol 2008; 58(2): 185-206.
- Chaby G, Senet P, Vaneau M, Martel P, Guillaume JC, Meaume S, et al. Dressings for acute and chronic wounds: A systematic review. Arch Dermatol 2007; 143(10): 1297-304.
- Edwards R, Harding KG. Bacteria and wound healing.Curr Opin Infect Dis 2004; 17(2): 91-96.
- Martin JM, Zenilman JM, Lazarus GS. Molecular microbiology: new dimensions for cutaneous biology and wound healing. J Invest Dermatol 2010; 130(1): 38-48.
- Sibbald RG, Orsted H, Schultz GS, Coutts P, Keast D. Preparing the wound bed 2003: focus on infection and inflammation. Ostomy Wound Manage 2003; 49(11):23-51.
- Howell-Jones RS, Wilson MJ, Hill KE, Howard AJ, Price PE, Thomas DW. A review of the microbiology, antibiotic usage and resistance in chronic skin wounds. J Antimicrob Chemother 2005; 55(2): 143-149.
- Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Applied and environmental microbiology 2008; 74(8): 2461-2470.
- Woo PCY, Lau, S.K.P., Teng, J.L.L., Tse, H., & Yuen,K.-Y. The and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clini Microbiol Infect 2008; 14: 908-9034.
- Harris KA, Hartley JC. Development of broad-range 16S rDNA PCR for use in the routine diagnostic clinical microbiology service. Journal of Medical Microbiology 2003; 52(8): 685-691.
- Wicke C, Bachinger A, Coerper S, Beckert S, Witte MB, Konigsrainer A. Aging influences wound healing in patients with chronic lower extremity wounds treated in a specialized Wound Care Center. Wound Repair Regen 2009;1 7(1): 25-33.
- Zhang X, Sarkar K, Rey S, Sebastian R, Andrikopou- lou E, Marti GP, et al. Aging impairs the mobilization and homing of bone marrow-derived angiogenic cells to burn wounds. J Mol Med (Berl) 2011; 89(10): 985- 95.
- Harker J. Wound healing complications associated with lower limb amputation2006. Available from: http://- www.worldwidewounds.com/2006/september/- Harker/Wound-Healing-Complications-Limb- Amputation.html.
- Swift ME, Burns AL, Gray KL, DiPietro LA. Age- related alterations in the inflammatory response to dermal injury. J Invest Dermatol 2001;117(5):1027-35.
- Athanasopoulos AN, Economopoulou M, Orlova VV, Sobke A, Schneider D, Weber H, et al. The extracellular adherence protein (Eap) of Staphyloco- ccus aureus inhibits wound healing by interfering with host defense and repair mechanisms. Blood. 2006; 107(7): 2720-2727.
- Edwards AM, Bowden MG, Brown EL, Laabei M, Massey RC. Staphylococcus aureus extracellular adherence protein triggers TNFalpha release, promo- ting attachment to endothelial cells via protein A. PLoS One 2012; 7(8): e43046.
- Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral JP, Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000; 38(10): 3623-30.
- Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007; 45(9): 2761-2764.