Research Article - Biomedical Research (2016) Computational Life Sciences and Smarter Technological Advancement
A novel approach for classification and clustering of biomedical citations
Parthasarathy G1*, Tomar DC21Department of Computer Science, Sathyabama University, Chennai, India
2Department of Information Technology, Jerusalem College of Engineering, Chennai, India
Accepted on April 05, 2016
Abstract
Citation refers the information of a published paper with its author and publication details. It is used by various authors for referring the research works published in other research articles. Citations play a crucial role in several scientific publications digital libraries (DLs), like Cite Seer, arXiv e-Print, DBLP, and Google Scholar. Users usually use citations to seek out data of interest in DLs, while researchers relay on citations to see the impact of a specific article. Citation mining is the area where in the citation databases are mined for performing various mining tasks such as classification and clustering to retrieve citations efficiently and accurately. Citations have additionally been used as auxiliary support in information retrieval tasks. Citation classification is the process of classifying the citation data by means of topic, author, paper name, and journal category. Clustering involves the categorization of papers based on content similarity or functional similarity. At present the size of databases in the web is massive hence the quantity of records in a dataset will vary from some thousands to thousands of millions. Authors or scholars are spending their precious time in searching the papers especially in bio medical field. So to provide more accurate retrieval of biomedical citations we have proposed a citation mining system with a combined approach of clustering. Our experiments conducted with the citations from the web database shows an effective retrieval of biomedical citations.
Keywords
Citation crawler, Citation mark-up language, Classification, Hierarchical clustering, Citation database.
Introduction
Citation is the reference of a published research work, referred by other authors. Citations are available in various forms in the web, called as citation databases. Citation databases represent the citation data of various journals. Citation mining involves the process of identifying the pattern of information present in the citation databases. Citation mining tasks are highly helpful for performing the extraction and analysis of citation data. Classification and clustering are the prominent mining tasks for performing mining on huge data. The classification of citations involves the process of splitting the large citation databases into many classified citation types. Clustering is the process of grouping similar data items into a single group. In general, clustering algorithms are classified into two categories which are hierarchical and non-hierarchical. Most of the hierarchical clustering algorithms make use of tree kind of cluster generation process. Non-hierarchical algorithms initially select the seeds and perform the clustering process by assigning the items into the clusters depending on the seeds. Dramatic increase within the range of educational publications has led to growing demand for economical organization of resources to fulfil researcher’s needs. Hence there is a necessity in adapting to a system which is efficient in retrieving citations. We proposed a citation mining system for powerful retrieval of bio medical citations and also to obtain precision in the results.
Related work
Citation analysis have witnessed a huge progress in various aspects of citation analysis tasks such as citation classification, citation clustering, citation database analysis, citation ranking. Most of the citation processing systems have used the frequencies of citations as a measure for ranking the journals. The early stage experiments in citation analysis evolved with citation indexing with the use of citation count measures from different journal publications. The advancement in the development of web technologies owed a way for spreading information across various places. But much of the information is not accessible to all the users. The available digital library information and ways to access those information efficiently are not enough satisfactory. If citations are easily accessible then indexing the citations provide lot of ways for analysing the citations and evaluating the journals. The citation indices maintained in the online repositories help to maintain scientific data for improving the effectiveness of the information access [1]. Mostly, all the research articles, journal and theses are made available in online. The researchers can easily make use of those papers by referring the idea or works discussed and implemented. Many times, the researchers face challenges for efficiently accessing that information. Earlier, the citations were represented using the tree structure and the various query mechanisms used lead to complex problems in accessing the data. Many citation related tasks have become a non-trivial one such as Topic based paper retrieval, exploring new research areas, retrieving semantically relevant papers and getting a specific paper in an area. This is addressed with the mapping of the citation retrieval task into a partitioning process. Each and every citation is mapped to a graph through reference links of papers. The citations are not evenly connected in the citation graph. The citation graph is sometimes a highly connected one with various links. Each connection in the graph helps to find topics, related topic contents and main citations. Since all these tasks are performed with efficient automation, the manual search process is significantly reduced and the results are more accurate [2]. Citation networks and scientific collaboration networks are in the centre of attention, nowadays. It is important that the compactness of the general network where how the metric can be used in citation analysis with citation collaboration network [3]. Citation analysis mainly involves the process of analysing the citation based on various citation databases. Scopus, Web of science and Google Scholar are the citation indexing system. There are various measures to compute the similarity between rankings indexed by different publications by sorting the citation count. There were more resemblances identified between rankings of Scopus and Web of science databases [4]. A research work on citation analysis used the topic information of 39 papers. The statistical techniques between these citation links allowed the discovery of most closed link among documents [5]. Clustering is the categorization of data items in an unsupervised manner. The clustering process has been used in different areas by various researchers. It is mainly helpful in data analysis process. There are various clustering algorithms available to many different applications. Clustering is used in various applications which are segmentation of images, recognition of objects and information retrieval [6]. A knowledge transfer model has been proposed using co-citation analysis. The model helps to analyse forward citations and backward citations for patent and non-patent data respectively. The co-citation clustering of the cited references considered as the bibliographical knowledge sources. The result of the analysis has shown that knowledge sources have evolved reasonably and the links have been established between the cited references. [7]. The users are spending their precious time in searching the papers. Thus, there a need arises for search engine optimization and precision of the results. The topics of programming, databases and operating systems have been used for the classification. The initial work of classification has used text mining techniques for searching the documents with the natural language represented in the best words to get the knowledge from the database. The classification uses the same search engine to identify the correct cluster from which the result can be obtained very efficiently [8]. It is necessary to use parser for parsing the paper content or components of citations available in any format. The parser has to capture the structural properties from semi structured format and those properties are transformed into a template having various properties used in structuring the citations. The structural properties of any citation include any kind of notation, even a small punctuation or any local structure at various fields of a citation. It is important to use encoding scheme with reserved words that is automatically trained for the data set to represent various unique symbol. A sufficient number of encoding templates are built, and then sequence alignment software is applied to check the matching of the input citation query with the sequencing templates [9]. Citation Analysis is the process of examination of the patterns, frequency graphs, and data in publications and articles. The impact of any research paper can be identified by computing the number of citations, the particular journal or paper received. Many researchers refer concepts or ideas from various papers and they quote the final outcome of the paper in their discussion. But it doesn’t mean that all the referring papers or authors would be criticizing the paper referred with positive words. Sometimes if the referring paper has made sensible results when put next to the referred paper then clearly the authors don't provide abundant positive comments in their discussion. In order to address this problem, the sentiment analysis can be performed to rate the citations based on the polarity level. The sentence parser is used to identify the adjectives what the authors used in their discussion about the cited paper. Then the determined adjectives can be assigned a score to distinguish it from positive and negative. In between, if any paper is identified without clearly mentioning the cited paper with identifiable adjective terms then it is named as unknown or neutral. Based on the computed polarity score, citations are ranked. The ranking process helps to project the citations which have cited with high positivity content in their discussion [10].
A Bayesian multi-causal model is used to compute the similarity between data items with the use of difference between the feature dimensions. The name of the author is given as the input query to the system. The queries have been given as input to the bibliographic search engines. The results are clustered with respect to the feature dimensions such as authors, title, location of the publication etc. [11]. An adapted leader method has been presented by combining it with a congenial adapted agglomerative hierarchical method. A cluster representative is considered as the leader. This method has been illustrated on the citation data of US patents. The distributed citations of patents have given better shape to output units [12]. The text based clustering techniques have been compared with the link based clustering techniques which determine the similarity based on the links represented with citing and cited references as in-links and out-links respectively. The citation context has combined along with the vocabularies in full text document. It has given a promising result in clustering journal articles. This kind of document representation strategy used with the clustering algorithm has outperformed the full text clustering method and the link based clustering techniques [13]. The challenging problem in citation networks is that the modelling of the high clustering. The existing studies have indicated that the better way to model the high clustering is the strategy of copying the references of the neighbour paper as its own references. A new method for high clustering has been proposed and it has achieved better cluster models with co-citation clusters in incorporating citation networks in multidisciplinary areas [14]. A system has been proposed for presenting information retrieval results in biomedical informatics. The goal of the system is to present the results with number of reduced citations with important citations in each group as prioritized. This text mining system performs automatic document clustering and ranking [15]. Multi-type data items with various types of relations are omnipresent especially in bibliographic networks. These networks are mixed information networks. But the research in the clustering is very limited. The algorithm has been evaluated with the help of data set crawled from ACM digital library [16]. A citation data clustering has been proposed to categorize the citation using the author names uniquely. The clustering approach is based on a probabilistic model which incorporates naïve bayes model and it is the extension of naïve bayes [17]. The performance of unsupervised clustering can be significantly improved by using the textual content with the graph structure of citations. Fisher’s inverse chi-square based hybrid clustering has been used. HITS and PageRank algorithms are used for representing the publication is each citation [18]. The connectivity analysis of linked documents provides significant details regarding the structure of document space for unsupervised learning activities. An information theoretic approach has been presented to measure the significance of individual words of the link between the documents. [19]. Generally, clustering algorithms make use of probabilistic based approaches. Though these probabilistic approaches are more flexible for providing better clustering, those approaches are very slower in cluster generation process. A fast, scalable clustering approach has been proposed to provide better clusters and also these clusters are very useful in modelling the citation data [20].
An unsupervised learning technique called k-way clustering for disambiguating citation author names. The technique has used three attributes of citations which are name of the co-authors, title of the papers, and location of the publications [21]. A comparative study has been performed to investigate different types of weighted citation networks for finding emerging research areas. The citation patterns containing citation, cocitation and coupling of bibliographies have been tested in three various domains [22]. The citation networks include the ideas of citation data from academics. The single linkage hierarchical clustering algorithm has been used for efficient clustering on large citation data. The distance measure clustering algorithms produce more and accurate clustering on citation data [23]. Our proposed system has further enhancement in efficiency among all the existent systems, thereby assuring supremacy in the process of acquiring biomedical citations.
Proposed work
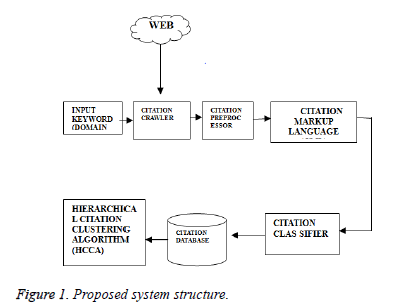
The efficient retrieval of biomedical papers from large source database in web becomes a crucial process. Hence the requirement to evolve to better system has proliferated. We have proposed a system for well-organized retrieval of biomedical citations based on certain algorithms. The citation mining architecture consists of the citation crawler, citation pre-processor, citation mark-up language, citation database, and hierarchical citation cluster and citation classifiers. Citation Crawler accepts the input keyword as seeds and extracts the papers from the web using the factor citation count. The downloaded PDF documents are reformed into text format for the ease of extraction process. Citation Preprocessor discards the citations based on two factors namely Citation count and Citation components. When a citation has minimum citation count and contains less than the required number of citation components, then the citation is rejected. Citation mark-up language receives citation elements in different formats. The received citations are converted into a common format (XML) type which makes it useful in processing. Citation Classifiers classify the citations based on the cited content as positive, negative, neutral and undefined with the use of classifiers. The classified citations are stored in the Citation Database and the hierarchical clustering algorithms are implied on it to obtain specified cluster as a result (Figure 1).
Citation crawler
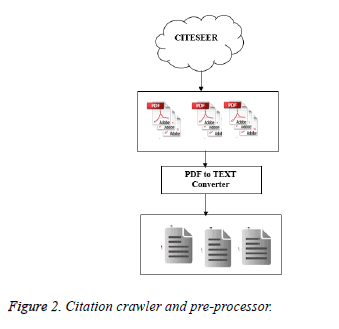
Crawler is a web spider, used to collect the citations from citation repositories like Google Scholar, Web of Science, and Cite seer. It uses the input domain keywords for collecting the citations. The domain terms are the keywords of specific technological domains. These keywords are given as the input seeds for gathering the citations. The crawler first gets the input seeds and it looks for the citation repositories and collects the citation information for the papers which are having maximum citation count. The citation count threshold is set to refine the citations that meet the threshold criteria. Figure 2 shows the citation pre-processor with citation crawler. The papers are downloaded from Cite seer databases and the collected data are organized. The PDF to text converter converts the downloaded PDF documents into plain text documents for ease of citation extraction. In addition to this,documents and their citations organized into a manageable data structure. The citation extractor extracts and formulates citations from each document and metadata.
Citation pre-processor
Generally, the collected citations consist of the citation components which are name, title, journal name, volume, issue, year, and page numbers. The pre-processor helps to refine the collected citations. It involves the process of removing the citations that have less citation count and citations not having all the citation components.
Citation mark-up language
The pre-processed citations are stored in an xml file that contains the citation elements as citation, paper name, author name, paper title, volume, issue, year, page number. These citation components are written in the xml file for all the collected citation with the mark-up tags. Since the citations are in different format, the collected components are written in the xml file and it provides a standard form for storing the citations.
Citation classifiers
Citation classification is the process of classifying the citations into different types based on the components of the citations. Most of the citation classification methods just classified the citations based on the author name, paper title, journal name for finding out the popularity of the cited author or reputation of the journal. No digital libraries distinguished the differences between the types of citations in the citation indexes. Most of the citations are mentioned just since other papers have referred it. None of the techniques have considered the citation based on whether it has been referred as positive or negative cited content. The classifiers perform the classification based on the cited content in the citing papers. The cited content of the papers have been collected and the intention of the citing paper that whether it has been referred with positive or negative content.
Citation database
The citation database consists of the citations that have been classified as positive or negative or neutral or undefined. The paper which just incorporated the concept from the citing paper is considered as the neutrally cited content. The citation database consists of 168870 citations collected from cite seer. Then the collected citations have been formatted and store with the various citation components. These citations are then used to perform the citation clustering with the labelled citations such as positive, negative and neutral.
Proposed algorithm
Algorithm steps
Input: domain keywords
Output: retrieved cited content
Step 1: Accepts input as domain keywords to the Citation Crawler.
Step 2: Retrieve the Citations from the web by the Crawler based on the Keywords specified.
Step 3: The pre-processor rejects the citation with low citation count and with insufficient citation components.
Step 4: The citation Mark-up language converts the citation components to a PDF format.
Step 5: Classification of citations as positive, negative, neutral and undefined with the use of citation classifiers.
Step 6: Storing of classified citations in the citation database.
Step 7: Calculating the Threshold value:
d (Ci, Cj)= min (i, j) where i belongs to Ci and j belongs to Cj
d (Ci, Cj)= max (i, j) where i belongs to Ci and j belongs to Cj.
Step 8: Clustering process is continued until when the distances between the clusters exceed the threshold value.
Experimentation and Results
Input keyword (Domain terms)
The web crawler is configured to recursively retrieve the websites related to the given input seeds. The input keywords are the key terms used to search the biomedical research articles in the digital libraries such as IEEE explore ACM digital libraries, Science direct, Google scholar and etc. The key terms specify the terms that used to represent the domain Biomedical. For example, the keyword Tumour [24], Pathology [25], Diabetes [26] etc. are used to retrieve the research papers related to the domain Biomedical. So Table 1 shows the keywords and the number of citations that have been collected. Once the documents were downloaded, the PDFs need to be gathered together and converted into text. For this process many PDF to text converter tools such as GHOSTVIEW, INTRAPDF, PDFBOX, and PDF Text Stream are reviewed. PDFBOX is more accurate and fast in the conversion process. Since Performance of PDFBOX is better, we use PDF Box engine from Apache for conversion and created a script to perform this operation. The total number of downloaded papers was around 3786. The size of the collection is about 3 GB, this might not seem a large amount however, and it probably explains the necessity of careful crawl and downloads. The following Table shows the results of the number of extracted citations and papers.
| Keyword | Number of papers | Number of citations |
|---|---|---|
| Chemotherapy | 446 | 20516 |
| Tumour | 570 | 39900 |
| Anatomy | 327 | 8829 |
| Neurology | 189 | 16821 |
| Histopathology | 130 | 3900 |
| Pathology | 540 | 35886 |
| Obstetrics | 229 | 6641 |
| Gynaecology | 639 | 24921 |
| Histology | 716 | 11456 |
| TOTAL | 168870 | |
Table 1. Input key terms for collecting citations from digital libraries.
Pre-processed citation collection
The total numbers of citations that have been collected from the digital libraries are 168870. These citations have been invoked to extract the citation components such as author name, paper title, journal name, volume number, issue number, year of publication and page number. Let A be the number of true positive, B be the number of false negative, and C be the number of false positive. The formulas (1), (2), (3) are used for computing the precision, recall, and F-measure.
Precision = A/(A+C) → (1)
Recall = A/(A+B) → (2)
F-Measure = (2*Precision*Recall)/(Precision+Recall) → (3)
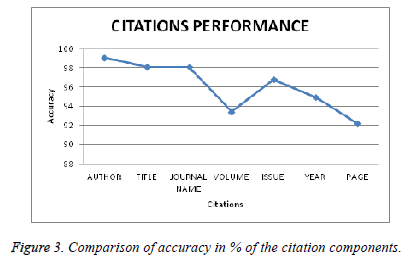
Table 2 shows the precision, recall, f-measure and accuracy of the citation components extracted from the citations. For example, the precision 99.31 of the citation component author represents that 99.31% of the author names from the citations have been extracted correctly.
| Precision | Recall | F-Measure | Accuracy | |
|---|---|---|---|---|
| Author | 99.31 | 99.5 | 99.41 | 99.1 |
| Title | 99.37 | 99.33 | 99.35 | 98.13 |
| Journal volume | 99.65 | 98.85 | 99.25 | 98.1 |
| Volume | 99.04 | 94.79 | 96.87 | 93.38 |
| Issue | 98.14 | 98.14 | 98.14 | 96.8 |
| Year | 99.54 | 96.9 | 98.21 | 94.93 |
| Page | 99.12 | 92.8 | 95.586 | 92.2 |
Table 2. Pre-processed results of the citations.
From the above graph we find that the citation component Author name has higher accuracy than the remaining citation components. This explains us among all the other components, author names of the citations are extracted more accurately.
Citation mark-up language for storing the preprocessed citations
Citation mark-up language (CML) is the xml based language which helps to represent the citation components of the extracted citations. The citation components that include author name, paper title, journal/conference name, volume, issue number, year and page number. The CML contains the citation representation of various papers.
Figure 4 gives the representation of CML for two of the citations extracted from tweet mining related papers.
Citation classifiers
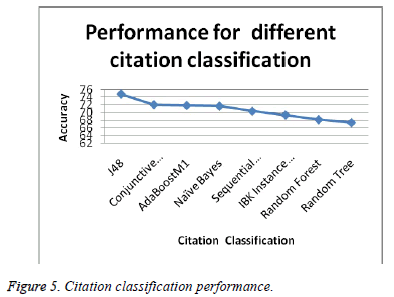
Citation classifiers are the standard data mining classifiers. Few of the classifiers that have been used for performing the citation classification are J48, Conjunctive rule, AdaboostM1, Naive Bayes, Sequential Minimal Optimization, IBK instance based, Random Forest and Random Tree. Table 3 shows the comparison between the classification results of the citations evaluated (Figure 5).
| Training citations (33774)' Testing citations (168870) | Total citations | Positive citations | Negative citations | Neutral citation | Undefined citations | Accuracy |
|---|---|---|---|---|---|---|
| J48 | 168870 | 126270 | 1350 | 5040 | 36210 | 74.78 |
| Conjunctive rule | 168870 | 121620 | 1410 | 4920 | 40920 | '2.03 |
| Ada boostMI | 168870 | 121260 | 1380 | 5040 | 41190 | '1.82 |
| Naive bases | 168870 | 120990 | 1380 | 3510 | 42990 | '1.65 |
| Sequential Minimal Optimization (SMO) | 168870 | 118770 | 1170 | 5010 | 43920 | '033 |
| IBIC Instance based | 168870 | 117090 | 1440 | 3960 | 46380 | 6934 |
| Random forest | 168870 | 116490 | 1440 | 5040 | 45900 | 68.11 |
| Random tree | 168870 | 114180 | 1440 | 3780 | 49470 | 6733 |
Table 3. Comparison of different classifiers results.
The corresponding graph for the different classifications mentioned above is given as under.
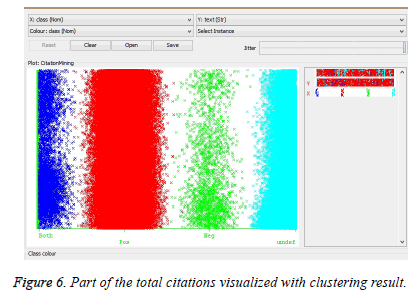
We can conclude from the above graph that J48 Classification has accuracy higher than the other classification. This higher accuracy establishes J48 with better performance. In our experiments, we used a dataset consisting of papers from biomedical fields. The data were drawn from Cite Seer, a digital library of papers from conferences and journals in bio medical fields. Cite Seer collects bio medical papers posted on the Internet as well as by linking directly to publishers, conference sites and journals, and then parses these articles to find the citations and descriptive information in each paper. It has over 7, 00,000 indexed papers in its database. Table 4 shows the class-wise detailed accuracy of the classifiers with the polarity based classification. Most of the classifiers have evaluated the citation data with good precision and recall rates. Table 4 shows the class-wise detailed accuracy of the classifiers with the polarity based classification. Most of the classifiers have evaluated the citation data with good precision and recall rates. The visualized citation clusters have been shown in Figure 6 There are four types of classified citations formed namely positive, negative, neutral (both) and undefined based on the labelled citation data. One of the classified result contain majority of the citations, which is the positive citation type. It has been highlighted in red colour in the visualized citation cluster. Then much of the citations have been classified as undefined, since they don’t have the content of positive or negative words. It has been indicated in cyan colour in the visualized citation classification result. The negative citations have been highlighted with green colour. Cited content of the citations that provide both positive and negative content, is highlighted as blue colour in the citation plot.
| CLASSIFIERS | DETAILED ACCL1R ACV | POSITIVE | NEGATIVE | UNDEFINED | BOTH |
|---|---|---|---|---|---|
| 348 | Precision | 0.767 | 0 | 322 | 0 |
| Recall | 0.933 | 0 | 0.423 | 0 | |
| F-measure | 0.842 | 0 | 0.643 | 0 | |
| Conjunctive rule | Precision | 0.736 | 0 | 31 | 0 |
| Recall | 0.954 | 0 | 0.145 | 0 | |
| F-measure | 0.831 | 0 | 226 | 0 | |
| AdaBoosM1 | Precision | 0.718 | 0 | 0 | 0 |
| Recall | 1 | 0 | 0 | 0 | |
| F-measure | 0.836 | 0 | 0 | 0 | |
| Nana Bayes | Precision | 0.718 | 0 | 0 | 0 |
| Recall | 1 | 0 | 0 | 0 | |
| F-measure | 0.836 | 0 | 0 | 0 | |
| SMO | Precision | 0.718 | 0 | 0 | 0 |
| Recall | 1 | 0 | 0 | 0 | |
| F-measure | 0.836 | 0 | 0 | 0 | |
| LEK Instance base | Precision | 0.773 | 0 | 536 | 0.04 |
| Recall | 0.816 | 0 | 0.407 | 0 | |
| F-measure | 0.799 | 0 | 0.463 | 0.05 | |
| Random forest | Precision | 0.785 | 0 | 474 | 0.05 |
| Recall | 0.78 | 0 | 0.467 | 0.05 | |
| F-measure | 0.787 | 0 | 471 | 0.05 | |
| Random tree | Precision | 0.785 | 0 | 0.469 | 0.04 |
| Recall | 0.782 | 0 | 0.469 | 0.04 | |
| F-measure | 0.783 | 0 | 0.469 | 0.05 |
Table 4. Class-wise detailed accuracy.
Combined approach
Clustering is the process of grouping similar data into one category. Citation clustering performs the categorization of the citations into different categories based on the classification results. That is, the citation data that already have been labelled as positive, negative, neutral and undefined are used for clustering. Based on these, the citations are clustered using Hierarchical and Non hierarchal algorithms. In order to make clusters combined (for agglomerative), or wherever a cluster ought to be split (for divisive), a live of dissimilarity between sets of observations is needed. In most strategies of stratified agglomeration, this can be achieved by use of an acceptable metric (a live of distance between pairs of observations), and a linkage criterion that specifies the dissimilarity of sets as perform of the pair wise distances of observations within the sets. Pre specified numbers of clusters are not needed for Hierarchical clustering. Hierarchical clustering algorithms are either top-down or bottom-up.
1. Agglomerative Hierarchical clustering algorithm (AGNES)-Bottom up Approach
2. Divisive Hierarchical clustering algorithm (DIANA)-Top down Approach
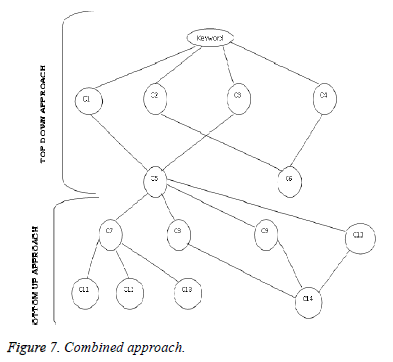
Agglomerative Hierarchical (Bottom up) clustering algorithm is defined as Agglomerative nesting. This algorithmic program works by grouping the information one by one on the idea of the nearest distance live of all the pair wise distance between the information purposes. This process continues until specified numbers of clusters are reached or there is only one cluster which contains all citations. The changes cannot be undone in this algorithm. It is immense to determine the exact number of clusters by the dendogram, this adds to an additional inconvenience in this method. Divisive Hierarchical clustering algorithm (Top down approach) is also known as divisive approach or deglomerative approach. The top-down clustering algorithm relies on a splitting technique. Thus, all documents are initially placed in one cluster, the top-down clustering algorithm proceeds by splitting clusters recursively until every cluster contains only citation. The complexity of this algorithm is greater than the agglomerative clustering. So we have proposed a method that combines both the top down and bottom up approach that makes it more efficient in clustering. Figure 7 show that the citations are clustered using combined approach. The citations classification made use of the cited contents of the citations that has been described with the sentiment polarity. The citations are classified as positive, negative, neutral and undefined using the classifiers. Further the clustering is applied in a way to group most similar citations in each classified category. The clustered citations are again classified as positive and neutral based on the sentimental polarity. The resultant citations are further classified. And the required citations are retrieved from the classified citations. This approach is more efficient and accurate in appropriate retrieval of biomedical citations.
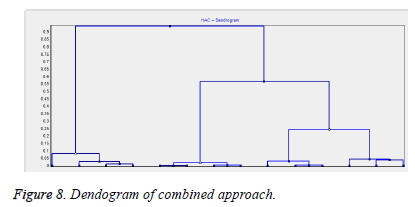
Figure 8 gives representation of the dendogram of the combined approach. Few citations have only been represented by the dendogram for interpretation of the clustering process.
Table 5 shows the clustering statistics that consists of the average, median, standard deviation, min*max, 1st* 3rd quartile range, skewness and kurtosis. The results show that cited content of citations is good alternative similarity identification, which provide further topical information on the source citation.
| Clustering Statistics | |
|---|---|
| Average | 73.5714 |
| Median | 72 |
| standard deviation | 6.5717 |
| Min*Max (full Range) | 5.3673 |
| 1st*3rd quartile (Range) | 69.00*80.00 |
| Skewness | 0.369 |
| Kurtosis | -0.8795 |
Table 5. Statistical measures of clustering data.
Especially they may contain useful synonymous and related terms that can upgrade the accuracy of the similarity calculation. We have proposed this system only for the purpose of retrieving citations from a particular field Biomedical. In future we can focus on the mining of citations on various trends. Table 6 shows the transformation of the data instances that have been transformed from continuous attribute values considered in the range of 50 to 100 to discrete set values. Since the polarity contained in each cited content represents with that range value. The count represents the number of instance with the range value specified in the values column and the percentage represents the total percentage of the instances. In Table 6, the values and percentages have been shown for the data instances.
| Values | Percent |
|---|---|
| X<66.1000 | 14.29 |
| 66.1000≤ x<68.2000 | 7.14 |
| 68.2000≤ x<70.3000 | 14.29 |
| 70.3000≤ x<=72.4000 | 21.43 |
| 72.4000≤ x<74.5000 | 0.00 |
| 74.5000≤ x<76.6000 | 14.29 |
| 76.6000≤ x78.7000 | 0.00 |
| 78.7000≤ x<80.0000 | 7.14 |
| 80.8000≤ x<82.9000 | 7.14 |
| x ≥ 82.9000 | 14.29 |
Table 6. Range of values and the corresponding percentage of instances.
Conclusion
As we have focused on the citation mining system in determining how efficacious a cited content of a biomedical citation representation with a series of experiments. Using citation polarity in the cited content of the citation, the clustering accuracy is improved. Since the cited content of the citation has already been identified, the clustering process looks for only the terms in the cited content rather than looking the entire paragraph of information of a cited paper. Therefore, it always reduces the cluster generation time. The results show that cited content of citations is good alternative similarity identification, which provide further topical information on the source citation. Especially they may contain useful synonymous and related terms that can upgrade the accuracy of the similarity calculation. We have proposed this system only for the purpose of retrieving citations from a particular field Biomedical. In future we can focus on the mining of citations on various trends.
References
- Jain K, Murty MN, Flynn PJ. Data clustering: a review, ACM Computing Survey 1999; 31: 264-323.
- Wang X, Zhao Y, Liu R, Jing Z. Knowledge-transfer analysis based on co-citation clustering. Scientometrics 2013; 97: 859-869.
- Jiang H, Lou W, Wang W. Three-Tier Clustering: An Online Citation Clustering System. Advances in Web-age information management 2001; 2118: 237-248.
- Kejzar N, Korenjak-Černe S, Batagelj V. Clustering of Distributions: A Case of Patent Citations. J Classi 2011; 28: 156-183.
- Aljaber B, Stokes N, Bailey J, Pei J. Document clustering of scientific texts using citation contexts. Info Retri 2010; 13: 101-131.
- Fu-Xin R, Xue-Qi CH, Hua-WS. Modeling the clustering in citation networks. Stat Mech Appli 2012; 391: 3533-3539.
- Yongjing L, Wenyuan Li, Keke CH, Ying L. A Document Clustering and Ranking System for Exploring MEDLINE Citations. J Am Med Inform Assoc 2007; 14: 651-661.
- Xiaoran XU,Zhi HD.BibClus: A Clustering Algorithm of Bibliographic Networks by Message Passing on Center Linkage Structure. IEEE International Conference on Data Mining, 2011; 864-873.
- Tomonari M, Atsuhiro T, Jun A. Citation data clustering for author name disambiguation, ACM International conference on Scalable Information Systems, 2007; 1-8.
- Frizo J, Wolfgang G, Bart DM. Dynamic hybrid clustering of bioinformatics by incorporating text mining and citation analysis. ACM SIGKDD international conference on Knowledge discovery and data mining 2007; 360-369.
- Bolelli L, Ertekin S, Giles LC. Clustering Scientific Literature Using Sparse Citation Graph Analysis, Knowledge Discovery in Databases. Lect Not Comp Sci 2006; 4213: 30-41.
- Vasileios K, Lyle U, Rajeev A. Online Clustering and Citation Analysis Using Streemer. Publicly Accessible Penn database, 2009.
- Hui H, Hongyuan Z, Giles CL. Name disambiguation in author citations using a K-way spectral clustering method. ACM/IEEE-CS Joint Conference on Digital Libraries 2005; 334-343.
- Fujita K, Kajikawa Y, Mori J, Sakata I. Detecting research fronts using different types of weighted citation networks. International Conference on Technology Management for Emerging Technologies 2012; 267-275.
- Michael JB II, Daniel MK, Jonathan LZ, James HF. Distance measures for dynamic citation networks. Physica A 2010; 389: 4201-4208.
- Parthasarathy G, Tomar DC. Sentiment analyzer: Analysis of journal citations from citation databases. Confluence The Next Generation Information Technology Summit, 2014.
- Egghe L, Rousseau R. BRS-Compactness in Networks: Theoretical Considerations Related to Cohesion in Citation Graphs, Collaboration Networks and the Internet. Math Comp Mod 2003; 37: 879-899.
- Judit BI, Mark L, Ayelet L. Some measures for comparing citation databases. J Inform 2007; 1: 26-34.
- Chyan Y, Szu-Hui WU, Lee J. A study of collaborative product commerce by co-citation analysis and social network analysis.IEEE International Conference on Industrial Engineering and Engineering Management, 2007; 24: 209-213.
- Chen D, Chi HC, Jing D, Chun LD. Citation retrieval in digital libraries.IEEE International Conference on Systems, Man, and Cybernetics, 1999; 105-109.
- Salim M, Mourad T. Ranking marketing journals using the Google Scholar-based hg-index. J Inform 2010; 4: 107-117.
- EA Calvillo, A Padilla, J Muñoz, J Ponce, JT Fernandez. Searching research papers using clustering and text mining.International Conference on Electronics, Communications and Computing (CONIELECOMP), Cholula, 2013, 78-81.
- Lu Q, Harbin C, Xiao L, Ye Q. Investigating the impact of online word-of-mouth on hotel sales with panel data, International Conference on Management Science and Engineering, 2012; 3-9.
- Ambeth KDV, Vaishali V, Shweta B. Basic Study of the Human Foot. Biomed Pharmcol J 2015; 8: 435-444.
- Jaafar SM. Diagnostic Implication and Clinical Relevance of Ancillary Techniques in Clinical Pathology Practice.Clini Medi Ins Path 2016; 9: 5-11.
- Dittakarn B, Vorama K. Incidence of large for gestational age infants when gestational diabetes mellitus is diagnosed early and late in pregnancy. J Obste Gynae R 2016; 42: 273-278.