Research Article - International Journal of Pure and Applied Zoology (2017) Volume 5, Issue 1
MORPHOLOGICAL, MOLECULAR AND KARYOLOGICAL ANALYSIS OF CLINOSTOMUM SCHIZOTHORAXI KAW, 1950 (DIGENEA) FROM FISHES OF KASHMIR VALLEY, INDIA
Fayaz Ahmad1, Omer Mohi ud Din Sofi2, Tanveer A Sofi1*, Khalid Majid Fazili3 and Bashir A Sheikh11Department of Zoology, University of Kashmir, Srinagar, India
2SK University of Agricultural Sciences and Technology, Shuhama, Aluestang, India
3Department of Biotechnology, University of Kashmir, Srinagar, India
- *Corresponding Author:
- Tanveer A Sofi
Department of Zoology, University of Kashmir, Srinagar, India
E-mail: stanveer96@gmail.com
Received Date: 27 January 2017; Accepted Date: 08 February 2017; Published Date: 20 February 2017
Abstract
The size of the amplified product of Clinostomum schizothoraxi was found 544 bp long and the sequence obtained was submitted to GenBank and their accession number acquired was AF973619. This species also showed 47% G+C content which reflects that they are also stable at high temperature. Specimens of Clinostomum schizothoraxi was found in fish hosts of Carassius carassius, Schizothorax niger, S. esocinus, S. curvifrons and S. plagiostomus as is evident from the NJ phylogenetic tree, which means that there is some correlation between Monogenean and Digenean parasites as far as their hosts are concerned. Clinostomum schizothoraxi showed different chromosome number as well as chromosome morphology as compared to that of Monogeneans. It Possess 20 numbers of diploid chromosomes (2n=20). First pair of chromosomes is metacentric; pairs 2 & 3 are submetacentric; pairs 4, 5 & 6 are subtelocentric and pairs 7, 8, 9 and 10 are acrocentric. The smallest chromosome measured 5.88 μm and the largest 9.15μm. Number of chromosome arms (NF) are to be 26 in number; total haploid complement length (TCL) is 77.12 μm. Arm ratio of the complement ranges between 1.51–18.31 and the centromeric index ranges from 5.18–39.88. The karyotype formulla (K) can been written as 2n=20=1m+2sm+3t+4a. This species has been studied for the first time with respect to molecular and chromosome analysis.
Keywords
Clinostomum schizothoraxi; Fish; Molecular; GenBank; Chromosome
Introduction
Digenetic trematodes comprise an estimated 24000 species, many of which have yet to be described (Poulin and Morand 2004). The Clinostomidae Luhe, 1901 is a family of digeneans the members of which, at the adult stage, live in the oral cavity, pharynx, or oesophagus of fish eating birds, reptiles and occasionally mammals, including man. Among the four subfamilies listed by Kanev et al. (2002) is the Clinostominae Luhe, 1901, which comprises three genera infecting piscivorous birds, such as herons, cormorants and pelicans; Clinostomum Leidy, 1856 is the type genus. Due to the high degree of morphological variability within the same species, in the past, Clinostomum has been subjected to several taxonomic revisions. The application of a molecular approach in parallel to karyological study may be particularly important for the completion of the life-cycle and identification of those Clinostomum species described in the past only on the basis of morphological features of the metacercarial stage without any subsequent description of the relative adult stages from the definitive host. Species-level identification of most trematodes is based exclusively on adult morphology. Difficulties arise because they are small,soft-bodied, have few stable morphological characters, and are subject to host-induced phenotypic variation (Blankespoor 1974; Graczyk 1991; Perez-Ponce de Leon 1995). In this context, molecular markers and chromosomal study offer powerful and much-needed tools that have the potential to distinguish between morphologically similar species at any stage in their life cycle.
Materials and Methods
Parasite Material
Mature specimens of clinostomids were recovered from fish hosts of Carassius carassius, Schizothorax niger, S. esocinus, S. curvifrons and S. plagiostomus from Anchar, Manasbal, Wular, Dal lake, River Jhelum and River Sindh of Kashmir valley, preserved in 100% ethanol for genomic DNA extraction and stored at -20°C for good quality of DNA. For DNA extraction ethanol was removed from parasites as per the protocol given in methodology and as such specimens were air dried to remove ethanol. The resultant DNA was examined on 1.5% agrose-TAE gels, stained with ethidium bromide and visualized under UV light.
Extraction of DNA
Phenol-Chloroform Technique: The detailed protocol is as follows (Sambrook and Russell, 2001):
Wash the parasite material in PBS.
↓
Place in a 1.5 ml eppendorf tube (UV cross- linked), grind with a pestle (UV treated) and add 300 μl of NET buffer.
↓
Add 15 μl SDS and 6 μl Protinase-K (10 mg/ml); store at -20°C.
↓
Incubate homogenate at 37°C overnight.
↓
Add 400 μl of 25:24:1 phenol/chloroform/isoamyl alcohol (stored at 4°C); mix the contents gently (~2 minutes).
↓
Centrifuge at 12,000 rpm for 10 minutes.
↓
Pipette out the upper phase (~280 μl) into a fresh 1.5 ml tube.
↓
Add 300 μl of 24:1 chloroform/isoamyl alcohol (stored at room temp.) to the sample; mix gently (~2 minutes).
↓
Centrifuge at 10,000 rpm for 10 minutes.
↓
Remove upper aqueous phase to a fresh 1.5 ml tube.
↓
Add 1 ml of ice-cold 100% ethanol (stored at -2°C) to the aqueous solution; mix gently; store at -2°C for 1 h.
↓
Centrifuge at 10,000 rpm for 10 minutes (DNA will precipitate).
↓
Remove supernatant.
↓
Wash the pellet in 1 ml 70% ethanol at 10,000 rpm for 10 minutes.
↓
Tip out the alcohol after the spin.
↓
Dry DNA pellet; resuspended in mili-Q water, store at 4°C.
Karyology
Trematode specimens of of Clinostomum spp. were obtained from gills, gill cover, mouth cavity, eyes and fins of fish hosts collected from the water bodies of Kashmir. Middle part of each individual containing testes and ovary was used for spreading analysis according to Frydrychova and Marec (2002). Parasite tissue was placed into hypotonic solution (0.075 M KCl) for 15-20 minutes and then put into freshly prepared Carnoy fixative (ethanol/ chloroform/ acetic acid, 6:3:1) for 20-30 min. Fixed tissue pieces were transferred into a drop of 60% acetic acid on a slide and thorn by tungsten needles making cell suspension. It was overspread along the slide on a heating plate (45°C). Slides were dehydrated in an ethanol series (70%, 80%, 100%, 1 min each) and stored at −20°C until further use.
Measurements are based on all chromosomes from 10 best metaphase spreads of parasites. The terminology relating to centromere position follows that of Levan et al., 1964. A chromosome is metacentric (m) if the ci falls in the range of 37.5–50.0, submetacentric (sm) if 25.0–37.5, subtelocentric (st) if 12.5–25.0; acrocentric (a) if <12.5 and telocentric if 0. When the centromere position was on the borderline between two categories, both are listed.
Statistical Analysis
Parametric as well as the non-parametric tests were used for analyzing relative length of chromosomes. A computer program (Minitab for windows) was used for data analysis. The descriptive data was given as a mean ± standard deviation (SD). Chi-Square analysis was used to see the statistical significance in chromosome lengths. Pearson’s correlation was used to find correlation between different species of helminths. Correlation analysis of data was carried out by using SPSS 16.5 package programme. Student’s t-test was used to test the differences, which were considered to be significant when the p-value obtained was less than 0.05.
Results and Discussion
Morphological analysis
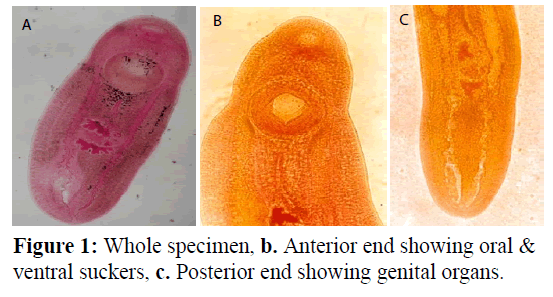
Clinostomum schizothoraxi Kaw, 1950 (Figure 1). Cysts enclosing metacercaria are spherical or broadly oval. Body of excysted worm elongated, linguiform, moderately fleshy with both ends rounded. Maximum width in or around region of gonads. Tegument aspinose. Oral sucker small, subterminal. Pharynx well developed, globular. Oesophagus practically absent. Intestinal caeca crenated on both sides behind level of acetabulum, inflated on one or both sides. Acetabulum large, muscular. Gonads immediately behind equatorial level of body. Shape of testes varies, being tandem, median, slightly lobed, morere or less triangular. Cirrus sac small, ovoid; lies on right side of anterior testis and ovary; enclosing a coiled seminal vesicle, pars prostatica and long and coiled ductus ejaculatorius entire, intertesticular, Genital pore on right side at about equatorial level of an anterior testis. Ovary generally, more or less globular, sometimes oval and irregular. Uteroduct, commencing at ootype, extends round anterior testis along its left margin and opens into uterine sac at about beginning of its posterior third of its length. Uterine sac long. Metraterm well developed.
Genital atrium nearly circular in outline. Genital pore submedian, lying on right side of middle of anterior testis. Vitelline follicles indistinct. Excretory pore subterminal; excretory bladder extending to level of blind ends of caeca. Comparative characteristics of this species as given by different workers are summarized in Table 1.
| Particulars | After Kaw, 1950 | After Dhar&Kharoo, 1986 | Fayaz&Chishti, 2000 | Bakshi, 1999 | Araet al., 2002 |
|---|---|---|---|---|---|
| Body length | 3.5-4.75 | 3.72-4.35 | 3.718 (3.2-4.2) | 4.2 (3.6-4.56) | 3.5–4.1 |
| Maximum width | 1.0-1.66 | 1.28-1.5 | 1.264 (1.0-1.48) | 0.956 (0.549-1.12) | 1.01- 1.09 |
| Oral sucker | 0.2-0.26 x 0.31-0.38 | 0.15-0.22 x 0.25-0.32 | 0.233 x 0.307 (0.176-0.35 x 0.208-0.353) | 0.224 x 0.356 (0.031-0.45 x 0.24-0.65) | 0.23 -0.25 x 0 .24-0.25 |
| Ventral sucker (Acetabulum) |
0.58-0.68 x 0.6-0.68 | ---------- | 0.524 x 0.584 (0.466-0.598 x 0.58-0.626) | 0.472 x 0.356 (0.031-0.45 x 0.24-065) | 0.6 x 0 .7 |
| Sucker length ratio | 1:2.73 | ---------- | 1:2.37 (1:1.48-2.84) | 1:2.105 | ---------- |
| Acetabulum distance from anterior end | ---------- | 0.42-0.50 | 0.565 (0.42-0.642) | ---------- | 0.49-0 .69 |
| Pharynx | ---------- | ---------- | 0.17 x 0.177 (0.16-0.19 x 0.16-0.192) | 0.59 x 0.43 (0.1-0.19 x 0.13-0.15) | 0.17 x 0 .19 |
| Anterior testis | 0.18-0.3 x 0.19-0.3 | 0.26-0.46 x 0.2-0.35 | 0.205 x 0.243 (0.176-0.257 x 0.176-0.305) | 0.284 x 0.223 (0.24-0.39 x 0.16-0.29) | ---------- |
| Posterior testis | 0.2-0.22 x 0.18-0.33 | 0.28-0.46 x 0.25-0.40 | 0.19-0.24 (0.15-0.224 x 0.144-0.385) | 0.234 x 0.302(0.21-0.29 x 0.019-0.17) | ---------- |
| Intertesticular distance | ---------- | ---------- | 0.19 (0.16-0.224) | ---------- | ---------- |
| Cirrus sac | 0.17-0.27 x 0.1-0.18 | ---------- | 0.22 x 0.088 (0.136-0.321 x 0.032-0.128) | 0.222 x 0.111(0.121-0.29 x 0.019-0.17) | ---------- |
| Ovary | 0.08-0.12 x 0.06-0.11 | ---------- | 0.076 x 0.085 (0.064-0.096 x 0.032-0.128) | 0.220 x 0.292 (0.10-0.29 x 0.24-0.35) | 0.141-0.152 x 0.72-0.079 |
| Uterine sac length | 0.5-0.6 | ---------- | 0.544 (0.433-0.691) | 0.762 (0.65-1.01) | 0.63-0.69 |
| Caecal termination to hind end distance | ---------- | ---------- | 0.11(0.032-0.2) | ---------- | 0.11-0.13 |
| Fish host | Schizothoraxniger; S. esocinus; Oreinussinuatus |
Cyprinuscarpiospicularis; Schizothoraxniger; S. esocinus; S. curvifrons; Nemachiluskashmirensis; OreinusplagiotomusandGlyptothoraxsp. |
Crassiuscrassius | Carassiuscarassius; Cyprinuscarpiocommunis | Schizothoraxesocinusand S. curvifrons |
| Locations of cysts | Beneath skin, base of fins, tail and gill covers. | Beneath skin, on fins, gills and gill covers. | Gills, Gill covers and eye sockets | Gills, Fins and Operculum | On fins, operculum, mouth cavity, Gill lamellae |
| Locality | River Jehlum, Kashmir | ---------- | Anchar and Mansbal lake | Dal and Manasbal lake | River Jhelum |
Table 1: Comparative characteristics (Measurements in mm) of Clinostomum schizothoraxi Kaw, 1950.
Remarks
The distinguishing features of the present metacercarial form are; Body with narrow forebody and broad hindbody; length ratio of oral sucker and acetabulum 1:2.71; Genital pore on the left side at the level of anterior border of anterior testis; testes lobed to a varying degree; ovary irregular in shape. When compared with metacercarial forms of known species of the genus Clinostomum, the present form stands near C. chrysichthys Dubois, 1930; C. gideoni Bhalerao, 1942 ; C. dasi Bhalerao, 1942 and C. piscidium Southwell and Prashad, 1918 in general disposition of gonads and shape of testes. However, the present form is found to be distinct in the position of the opening of uteroduct into the uterine sac. From C. dasi it further differs in the position of genital pore.
Molecular Analysis
PCR Amplification
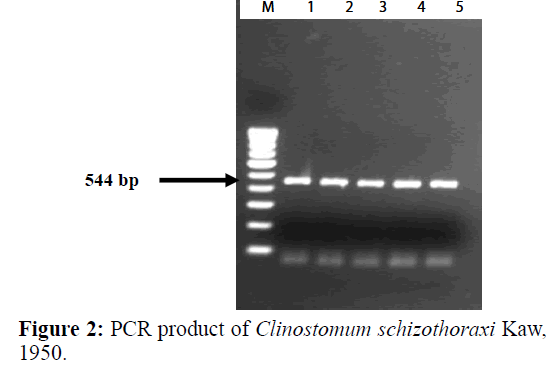
PCR amplification was carried out to amplify ITS region of the Digenean parasite (Table 3). The size of the amplified product was found to be 544 bp long (Figure 2). In BLAST search of the sequences, it showed maximum similarity with Clinostomum cutaneum Based on morphological characters, this species was identified as belonging to Clinostomum schizothoraxi. The present results of the molecular analysis confirmed the species identification of this parasite. Therefore, it can be assumed that the present form recovered from the fish host in Kashmir valley is Clinostomum schizothoraxi Kaw, 1950.
| Species | Host of adult from (Avian Hosts) | Host of metacercarial from (Fish Hosts) | Synonym |
|---|---|---|---|
| C. piscidium, SouthwelletPrashad, 1918 | Ardeolagrayii Exper. By Singh (1959) |
Trichogasterfasciatus; Nandusnandus |
C. complanatum, Ukoli (1966) C. marginatum, Agarwal (1959) |
| C. dasiBhalerao, 1942 | Bubulcus ibis and Ardeagrayii Exper. By Pandey (1966) |
Saccabranchusfossilis; Heteropneustesfossilis |
C. companatum, Ukoli (1966) |
| C. gideoniBhalerao, 1942 | ---------- | Barbussophare | C. complanatum, Ukoli (1966) |
| C. gideoniBhlerao, 1942 | ---------- | Unidentified fish | ---------- |
| C. anusi Wesley, 1944 | Xenorthynchusasiaticus | Unidentified fish | C. complanatum, Ukoli (1966) |
| C. sp. Srivastana, 1950 | Bubulcuscormandus; Nycticoraxnyticorax Exper. By Srivastava (1950) |
Channa (Ophiocepha) punctatus |
---------- |
| C. schizothoraxi, Kaw, 1950 | ---------- | Oreinussinuatus; Schizothoraxesocinus; S. niger |
---------- |
| C. deccanum, Jaiswal, 1957 | Ardeacinerea | ---------- | C. complanatum, (Rud. 1809); Ukoli (1966) |
| C. demiegrattae, Jaiswal, 1957 | Demiegrettaasha | ---------- | C. complanatum, Ukoli, (1966) |
| C. hydrabadensis, Jaiswal, 1957 | Ardeagravii | ---------- | C. complanatum, Ukoli, (1966) |
| C. macrostomum, Jaiswal, 1957 | Tilapia nilotica(Linn.) | Channa (Ophiocephalus) striatus | C. mastacembeli, Ukoli, (1966) |
| C. mastacembeli, Jaiswal, 1957 | ---------- | Mastacembelusarmatus | Syn. C. macrostomum, Ukoli, (1966) |
| C. singhi, Jaiswal, 1957 | Ardolagravii | ---------- | C. complanatum, Ukoli, (1966) |
| C. gignticum, Agarwal, 1959a | Bubulcus ibis and ArdeolagraviiExper. By Agarwal (1959a) |
Channa (Ophiocephalus) Punctatus; Bariliusbendelisis |
C. complanatum, Ukoli, (1966) |
| C. kalappah, Bhalerao, 1947 | (Mammalian Host) Accidentally – Cat Probably bird host in nature |
---------- | ---------- |
| C. attenuatum, Cort, 1913 | Ardea Herodias, Jaiswal (1957) Botaurus, Ardea – Hunter et Hunter (1934) |
(Amphebian Host) Ranapipens (U.S.A) R. tigrina – Cort (1913), Jaiswal (1957) |
C. marginatum (Wright, 1879 nec. Rud., 1809) Beer (1933) Valid – Ukoli (1966) |
| C. progonum, Jaiswal, 1957 | ---------- | Ranacyanophlyctis | C. complanatum, Ukoli (1966) |
Table 2: Species of Clinostomum described from Indian region.
| Digenea | Initial Denaturation | Denaturation for 40 cycles | Annealing | Extension | Final extension |
|---|---|---|---|---|---|
| Clinostomumschizothoraxi | 95oC for 10 minutes | 40 cycles at 95oC for 30 seconds | 52oC for 30 seconds | 72oC for 75 seconds | 72oC for 10 minutes |
Table 3: PCR assay of Digenea which were carried out in a thermocycler (Eppendorf Mastercycler Personal) under different conditions.
Nucleotide Sequences
The size of the nucleotide sequence was found to be 544 bp long. The sequence obtained were submitted to GenBank and their accession number acquired was AF973619 (Table 4). Sequences were compared with other sequences of digenean trematode species from GenBank. The consensus sequences analysed by BLAST search gave a 93.25% identity with C. cutaneum (GQ339114.1) and 85% with C. complanatum (FJ609420.1). The alignment of 28S rDNA sequences of Clinostomum schizothoraxi showed few differences between the species of other digeneans. The nucleotide sequences obtained shown in Figure 3 are raw sequences.
| S. No. | Digenean Species | Host | Accession No | Family | Base pairs | Authors | Country | Year |
|---|---|---|---|---|---|---|---|---|
| 1 | Clinostomumschizothoraxi Kaw, 1950* | Carassiuscarassius; Schizothoraxniger; Schizothoraxesocinus; Schizothoraxcurvifrons | AF973619 | Clinostomatidae | 544 bp | Present study | India | 2015 |
| 2 | Clinostomumcutaneum | Grey herons Ardea cinerea L. and Nile tilapiaOreochromisniloticusniloticus (L.) |
FJ609421 | Clinostomatidae | 4614 bp | Caffara and Stanzani | Kenya | 2010 |
| 3 | Clinostomumcutaneum | Ardeacinerea | GQ339114 | Clinostomatidae | 4637 bp | Gustinellietal., | Kenya | 2010 |
Table 4: Digenean trematode species used for molecular comparison of ITS DNA sequences along with their hosts, country and GenBank accession numbers for corresponding sequences (*Query sequence).
The present observation shows that Clinostomum schizothoraxi has 544 base pair length with 171 amino acids. This species is more stable at higher temperature with 47% G+C content (Table 5).
| Length | A | C | G | T | G+C |
|---|---|---|---|---|---|
| 544 bp | 127 | 112 | 148 | 157 | 47% |
| Total No. of Amino Acids | 171 | ||||
| Molecular Weight | 19250 Da | ||||
Table 5: Summary of base pairs and amino acids of Clinostomum schizothoraxi Kaw, 1950.
Pairwise Alignment
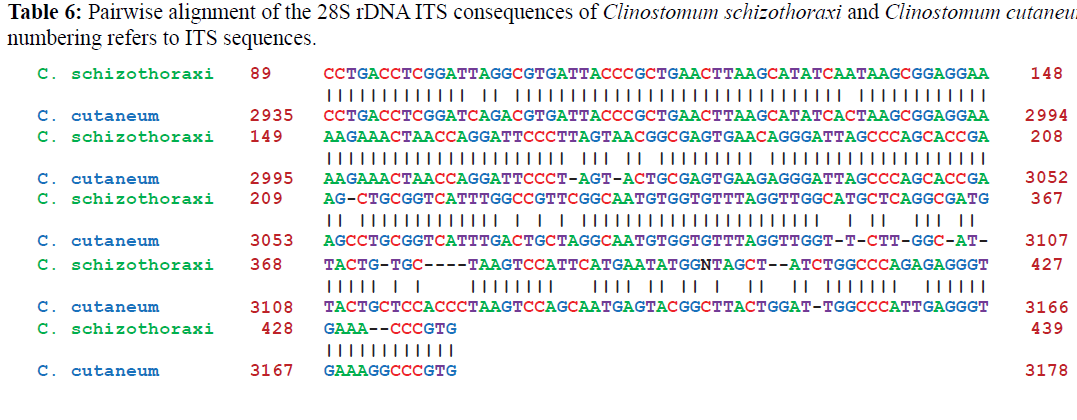
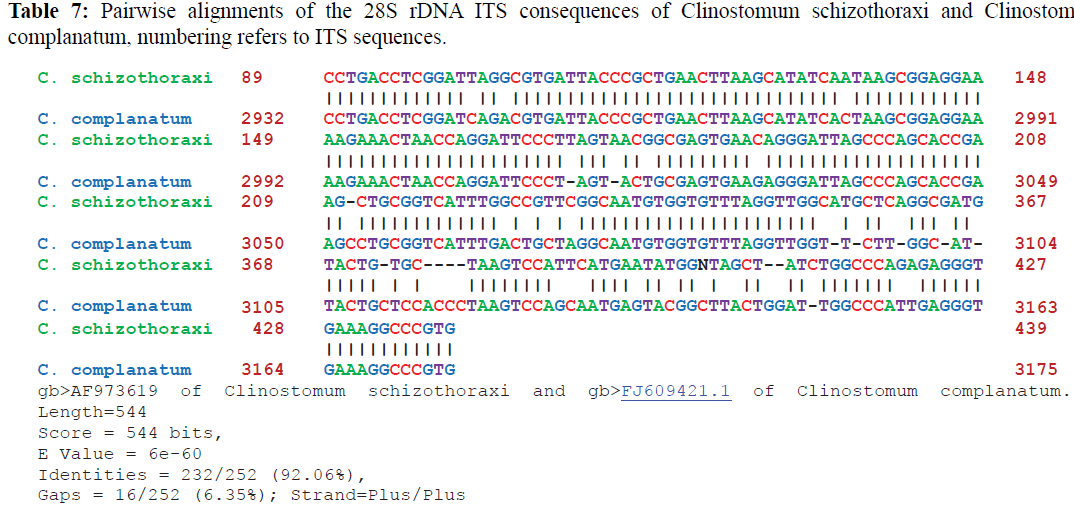
Pairwise alignments of Clinostomum schizothoraxi was made by using different softwares as discussed in materials and methods. Clinostomum schizothoraxi showed maximum similarity with those of Clinostomum cutaneum and Clinostomum complanatum as shown in the following Tables 6 and 7.
Construction of Phylgenetic Tree
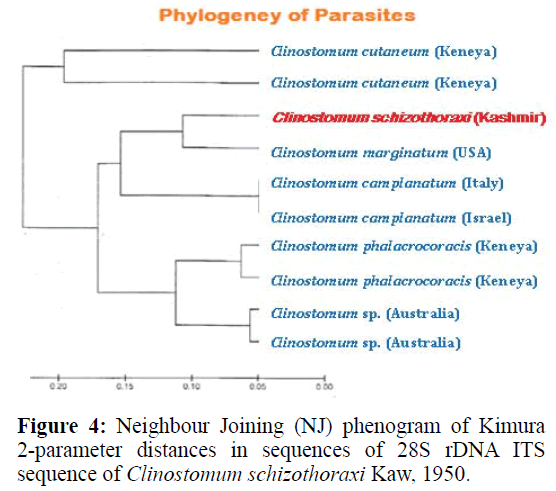
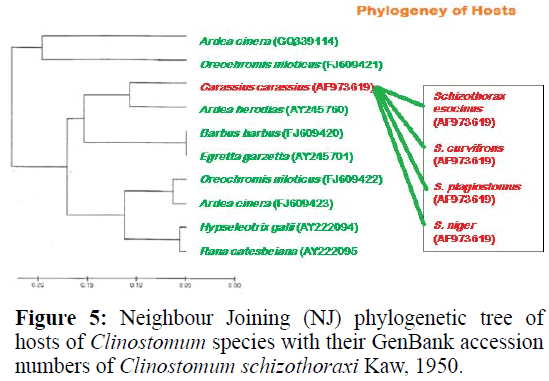
For phylogenetic analysis, Maximum likelihood (ML); Maximum Parsimony (MP) and Neighbour Joining (NJ) trees were constructed (Figurtes 4 and 5). The ITS sequences of Clinostomum schizothoraxi were compared with the sequences available for other Digenean parasites in the GeneBank database. The trees showed that the query ITS of Clinostomum schizothoraxi sequences stand close to of C. cutaneum.
It is clear from the Tables 6 and 7 that Clinostomum schizothoraxi with GeneBank accession number shows AF973619 shows 93.25% and 92.06% resemblance with that of Clinostomum cutaneum (GQ339114.1) and Clinostomum complanatum (FJ609421.1) with 16 gaps respectively. Out of 252 base pairs Clinostomum schizothoraxi, 235 & 232 base pairs match with that of Clinostomum cutaneum & Clinostomum complanatum respectively.
Discussion
Clinostomum schizothoraxi was first described for the first time by Kaw (1950) from Schizothorax niger, S. esocinus and Oreinus sinuatus from the River Jehlum and then by Dhar (1978) and Dhar and Kharoo (1986) from Cyprinus carpio specularis, S. curvifrons, Nemachilus kashmirensis, Oreinus plagiostomus and Glyptothorax sp. In order to contribute to the known molecular data on metacercarial form of Clinostomum species, we sequenced almost all of the 28S rDNA of Clinostomum schizothoraxi from Scizothorax and Carassius spp. collected from Kashmir valley. Comparison of the alignments obtained from sequences of the 28S rDNA genes and the internal transcribed spacer regions ITS1 and ITS2 of Clinostomum schizothoraxi with the other Digenean species sequences available in GenBank produced interesting results. The present observation showed that the alignment of the 28S rDNA exhibited few differences between the Clinostomum schizothoraxi and C. cutaneum, with distances of 6.75%. As described by Hills and Dixon (1991), this region is highly conserved and is characterized by a slow evolutionary rate, useful for evaluating ancient evolutionary events however; it may be useless in some cases for discriminating organisms at the species level but in the case of the present study, the alignment showed little differences between the species. Several authors (Hills & Dixon, 1991; Hershkovitz & Lewis, 1996; Coleman, 2003) described this gene as characterized by a high level of conservation similar to that of 18S rRNA gene, so it is used in phylogenetic studies (Troitsky & Bobrova, 1986; Troitsky et al., 1991; Suh et al., 1992). In addition, this coding region is too short 160 bp in Clinostomum species, to produce phylogenies across large time scale.
The internal transcribed spacers, ITS1 and ITS2, are relatively conserved regions within species or genera and have been used as markers in population genetic studies (Hillis & Dixon, 1991) and to explore species boundaries in digenean families (Nolan & Cribb, 2005). During the present study it was observed that ITS1 of C. schizothoraxi is characterized by the presence of tandem repeat units at the 50 end that provide the variability in the sequence composition at both the interspecific and intraspecific level. These repeats are also known in some digenetic trematode families, such as the Haematoloechidae, Mesometridae, Opecoelidae, Schistosomatidae, Strigeidae and Telorchiidae (Nolan & Cribb, 2005) and support the present observation. The ITS1 sequences of C. schizothoraxi were 544 bp long, and interspecific variation in the length of the ITS1 was observed, like the observations of others authors, such as van Herwerden et al. (1998) for Schistosoma spp. and Dvorak et al. (2002) for Trichobilharzia spp. The different lengths of ITS1 are due to the presence of the repeat elements (Luton et al., 1992; Kane & Rollinson, 1994; Kane et al., 1996), which supports our results. The ITS2 spacer is characterized by a lower degree of variation with a high degree of conservation at the species level. Usually, it does not contain repeat units, although Morgan & Blair (1995) found repeat units of ITS2 in the Echinostomatidae. It is also characterised by differences in length within and between families. In our sequences, we found different length i.e., 544 bp in C. schizothoraxi moreover, we did not observe intraspecific variations as described by some authors for other digeneans (Luton et al., 1992; Galazzo et al., 2002; Jousson & Bartoli, 2002). The 28S rDNA gene is longer and has more variations in the rate of evolution than the 18S rRNA. This gene has been used in phylogenetic studies (Olson et al., 2003). The alignment of the 28S rDNA sequences obtained showed very low differences between the sequences compared, particularly between C. schizothoraxi and C. cutaneum with only 17 nucleotide differences.
Even though sequencing DNA represents the primary approach of modern systematics, the combination with traditional techniques, such as the use of morphological parameters, is essential for describing parasites at the species level. In fact, as suggested by Nolan & Cribb (2005), the best way to approach species identification is to perform morphological descriptions on half the specimens and molecular analysis on the other half.
In the present study, we described for the first time the metacercarial form of C. schizothoraxi from cyprinid fishes of Kashmir valley, the definitive host of the parasite, combining a traditional morphological approach with molecular analyses. Besides sequencing the DNA of our parasite, we were also able to link the adult stage to the metacercarial stage found in Kashmir valley from the same environment, confirming the morphological observations. Following the suggestion of Matthews & Cribb (1998), who suggested the need for a reorganization of the genus Clinostomum using both morphological and molecular approaches, this study hopefully represents the first input towards a systematic revision of this complex group of parasites.
Karyological Analysis
The Clinostomidae Luhe, 1901 is a family of digeneans, the members of which live in the oral cavity, gills, gill covers, eye sockets, operculum, fins, and gill lamellae of fishes. Due to the high degree of morphological variability within the same species, Clinostomum has been subjected to several taxonomic revisions (Gustinelli et al., 2010). The application of a karyology in parallel to morphological study may be particularly important for the identification of Clinostomum species described in the past only on the basis of morphological features.
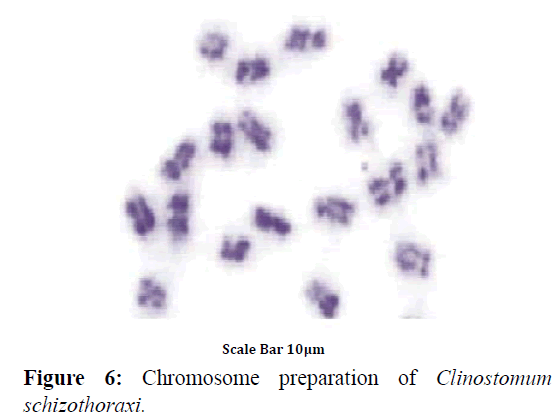
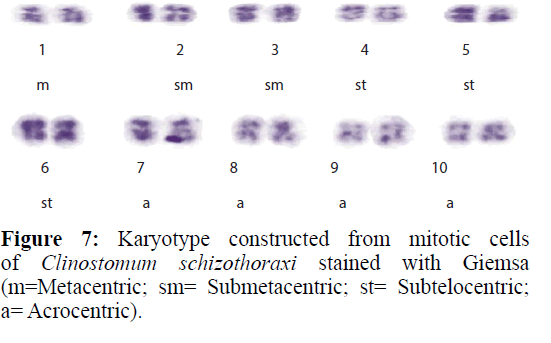
Colchicine treatment allowed us to examine enough number of metaphase plates with well-contracted mitotic chromosomes. In this way, the number and morphology was determined with high accuracy. A diploid complement of 2n=20 was found in 43 dividing cells of Clinostomum schizothoraxi (Figure 6) collected from Schizothorax and Carassius spp. The chromosomes are large; the smallest measured 5.88 μm and the largest 9.15 μm (Table 9).
| Description | Max score | Total score | Query cover | E value | Identity | Accession No. |
|---|---|---|---|---|---|---|
| Clinostomumcutaneum voucher 129/09 18S ribosomal RNA gene, internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence | 241 | 241 | 24% | 2e-59 | 93.25% | GQ339114.1 |
| Clinostomumcutaneum voucher 118/07 18S ribosomal RNA gene, internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence | 241 | 241 | 24% | 2e-59 | 92.06% | FJ609421.1 |
| Clinostomumcomplanatum voucher 297/02 18S ribosomal RNA gene, internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence | 241 | 241 | 24% | 2e-59 | 85% | FJ609420.1 |
| Clinostomumphalacrocoracis voucher 10/08 18S ribosomal RNA gene, internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence | 241 | 241 | 24% | 2e-59 | 82% | FJ609422.1 |
| Clinostomumphalacrocoracis voucher 16/08-2TA 18S ribosomal RNA gene, internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence | 233 | 233 | 24% | 4e-57 | 78% | FJ609423.1 |
Table 8: Sequences producing significant alignments in Clinostomum species.
| Chromosome pair number | Length of short arm (µm) ‘S’ | Length of long arm (µm) ‘L’ | Total Length/Absolute Length (µm) L+S | Arm Ratio (L/S) | Relative Length (%) |
Centromeric Index (ci) |
Classification | |
|---|---|---|---|---|---|---|---|---|
| 1 | 3.21 | 4.84 | 8.05 | 1.51 | 10.44 | 39.88 | Metacentric | |
| 2 | 2.13 | 4.11 | 6.24 | 1.93 | 8.09 | 34.13 | Submetacentric | T-Value = -32.64 P-Value = 0.020 P<0.05 |
| 3 | 2.00 | 3.88 | 5.88 | 1.94 | 7.62 | 34.01 | Submetacentric | |
| 4 | 1.54 | 6.63 | 8.17 | 4.31 | 10.59 | 18.85 | Subtelocentric | T-Value = -52.78 P-Value = 0.012; P<0.05 |
| 5 | 1.33 | 6.52 | 7.85 | 4.90 | 10.18 | 16.94 | Subtelocentric | |
| 6 | 1.00 | 5.43 | 6.43 | 5.43 | 8.34 | 15.55 | Subtelocentric | T-Value = -9.54 P-Value = 0.011 P<0.05 |
| 7 | 0.84 | 8.31 | 9.15 | 9.89 | 11.86 | 9.18 | Acrocentric | |
| 8 | 0.71 | 8.13 | 8.84 | 11.45 | 11.46 | 8.03 | Acrocentric | |
| 9 | 0.56 | 7.84 | 8.40 | 14.00 | 10.89 | 6.67 | Acrocentric | T-Value = -61.25 P-Value = 0.010 P<0.001 |
| 10 | 0.42 | 7.69 | 8.11 | 18.31 | 10.52 | 5.18 | Acrocentric | |
Table 9: Measurements and classification of chromosomes of Clinostomum schizothoraxi Kaw, 1950.
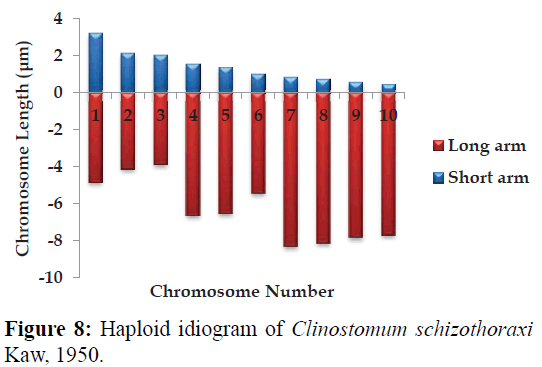
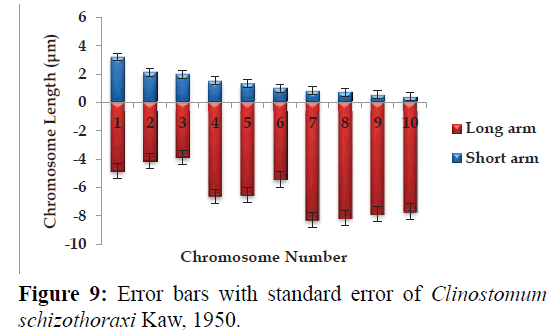
Karyotype (Figures 3 and 7) included one metacentric (no. 1); two submetacentric (no.s 2 and 3); three subtelocentric (no.s 4; 5 and 6) and four acrocentric (no.s 7, 8; 9 and 10) chromosome pair; the karyotype formula may be summarized as 2n=20=1m+2sm+3t+4a. Number of chromosome arms (NF) was 26; total haploid complement length (TCL) was 77.12 μm. Arm ratio of the complement ranges between 1.51–18.31 and the centromeric index ranges from 5.18–39.88 (Table 4). There is less chromosome length difference between 2 & 3; 4 & 5, 6, 7 & 8 and 9 &10 which are statistically less significant (P<0.05 ; Students T-test). On the basis of absolute length and centromeric position the chromosomes have been arranged in order of decreasing length in an ideogram (Figures 8 and 9) and the karyotype formula is: (K) 2n=20=1m+2sm+3t+4a.
Discussion
Digenean Trematodes are karyotypically conservative, and their karyotypes tend to have the same number and closely related gross chromosome morphology of the genus and family (or even higher) taxonomic level. Most of the karyologically studied members of Digenean families are Troglotrematidae, Plagiorchiidae, Telorchiidae, Prosthogonimidae, and Lecithodendriidae. Information on the cytogenetics of Clinostomidae trematodes is limited largely to the description of the chromosome numbers. Clinostomum schizothoraxi possess 10 haploid chromosomes in which first pair is metacentric; pairs 2 & 3 are submetacentric while pairs 4; 5 & 6 are subtelocentric and pairs 7; 8; 9 and 10 are acrocentric which were observed during the present investigation. Britt (1947) also reported 10 haploid chromosomes in various species of the family Clinostomidae. According to Britt (1947), the largest chromosome were observed in the family Allocreadiidae, where some pairs measures up to 8 μm in length and smallest in the families Clinostomidae and Plagiorchiidae, in case of the present study the chromosomes are medium sized; the smallest measured 5.88 μm and the largest 9.15 μm. His work was mainly concerned with collecting cytological evidences to find an evolutionary mechanism in Platyhelminthes. However, Short and Menzel (1959) concluded that morphological changes accompanying separation of the genera seem to have been the results of translocations, inversions, deletions, and changes in the number of smaller chromosomes of Digeneans. The present results on karyotype of Clinostomum schizothoraxi, 2n=20, are in agreement with Britt (1947) who described 20 chromosomes of Clinostomum marginatum. Despite limitations of the method used, Britt (1947) established that small chromosomes are characteristic of Clinostomidae species and our data confirm these findings. Mitotic chromosomes of Clinostomum schizothoraxi are medium sized, up to 9.15 μm. Notable workers like Cable (1931, 1974); Anderson (1935); Chen (1937); Rees (1937); Pennypacker (1936, 1940); Markell (1943); Ciordia (1949; 1950, 1956); Willmott (1950); Willey and Godman (1950); John (1956); Dhingra (1954a-c,1955a-c); Guildford (1955, 1961); Short, (1957) and Short and Menzel (1957, 1959); Sanderson (1959); Barton (1960); Greson (1958, 1964); Sharma et al. (1968); Saksena (1969); Reddy and Subramanyam (1973, 1975); Sharma & Nakhasi (1974) and Jha (1975) have observed interesting cytological variations regarding the chromosome number and morphology in the order Digeneans.
Changes in chromosome form in digeneans trematodes were believed most commonly from centric fusion, pericentric inversions, changes in the amount of heterochromatin and euchromatin, and through other chromosomal rearrangements (Grossman & Cain, 1981). Often a likely route for the evolution of chromosomes within a family or genus can be visualised; key indicators include: karyotypic variation between related species whose diploid complements differ (Barsiene & Grabda- Kazubska, 1988); atypically large chromosomes (Grossman & Cain, 1981); the relative lengths of chromosomes comprising the haploid genome (White, 1973); and the chromosome arm number (Mutafova, 1994). One of the most commonly observed evolutionary pathways occurring in the digenean genome results from a Robertsonian translocation.
Acknowledgements
The authors extend their thanks to the authorities of the Department of Zoology & Biotechnology, University of Kashmir for the facilities provided. TAS is also highly thankful to Prof. Fayaz Ahmad for giving valuable suggestions while compiling this paper.
References
- Anderson, М. G., 1935. Gametogenesis in the primary generation of a digenetic trematode, ProterometramacrostomaHorsfall, 1933. Trans. Amer. Micr. Soc. 54: 271-297.
- Ara, J., Khan, A. R. and Fayaz, A., 2002. First record of a PseudophyllideanCestodeBothriocephalus (Rudolphi; 1808) from fishes of Kashmir. Oriental. Sci. 5: 23-26.
- Bakshi, S. A., 1999. Fish parasites of some lakes of Kashmir with an analysis of seasonality of incidence and their maturation with regard to different ecological factors. Doctoral Thesis, University of Kashmir, India.
- Barsiene, J. V. and Grabda-Kazubska, B., 1988. A comparative study on chromosomes in plagiorchiidtrematodes. I. Karyotypes of Opisthioglypheranae (Frolich, 1791), Haplometracylindracea (Zeder, 1800) and Leptophallusnigrovenosus (Belingham, 1844). Acta.Parasitol. Polonica. 33: 249-257.
- Barton, P. R., 1960. Gametogenesis and fertilization in the frog lung fluke Haematoloechusmedioplexus Stafford (Trematoda: Plagiochiidae) J. Morph. 107: 93-122.
- Blankespoor, H. D., 1974. Host-induced variation in Plagiorchisnoblei Park, 1936 (Plagiorchiidae: Trematoda). Am. Midland Nat.92: 415-433.
- Britt, H. G. 1947. Chromosomes of Digenetic Trematodes. The Am Nat.81(799): 276-296.
- Cable, R. M., 1931. Studies on the germ-cell cycle of CryptocotylelinquaCreplin. I. Gametogenesis in the adult. Quarterly Journal of Microscopic Science74: 563-589.
- Cable, R. M., 1974. Phylogeny and taxonomy of the trematodes with reference to marine species. In W. A. Vernberg (edn.), Symbiosis in the Sea. Univ. South Carolina Press, Columbia, pp 173-193.
- Caffara, M. and Stanzani, E., 2010. First description of the adult stage of ClinostomumcutaneumPaperna, 1964 (Digenea: Clinostomidae) from grey herons Ardeacinerea L. and a redescription of the metacercaria from the Nile tilapia Oreochromisniloticusniloticus (L.) in Kenya. Syst. Parasitol. 76: 39‐51.
- Chen, P. D., 1937. The germ cell cycle in the trematode, Paragonimuskellicotti Ward. Trans. Amer. Micros. Soc. 56: 208-236.
- Ciordia, H., 1949. Cytological Study of Rhopaliasmacracanthus Chandler, 1932, a Trematode from the Opossum, Didelphisvirginiana. The Journal of Parasitology35: 417-422.
- Ciordia, H., 1950. The chromosomes of Notocotylusfilamentis Barker, 1915, a monostome from the muskrat (Fiberzibethicus). Trans. Amer. Micr. Soc. 69: 64-65.
- Ciordia, H., 1956. Cytological studies on the germ cell cycle of the trematode family Bucephalidae. Trans. Amer. Micr. Soc. 75: 103-116.
- Coleman, A. W., 2003. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends in Genetics19: 370-375.
- Dhar, R. L., 1978. Studies on the Helminth Parasites of Fishes of Jammu and Kashmir. Ph.D. Thesis, University of Kashmir, Srinagar,India.
- Dhar, R. L. and Kharoo, V. K., l986. Studies on trematode parasites of fishes-Genus Clinostomum Leidy, 1856 from freshwater fishes of Kashmir. Ind. J. Helminthol. 38: 74-78.
- Dhingra, O. P., 1954a. Taxonomic values of chromosomes and cytoplasmic inclusions in a digenetic trematode-Phyllodistomum spatula. Res. Bull. Panjab. Univer. Zool., 51: 101-109.
- Dhingra, O. P., 1954b. Gametogenesis and fertilization in Isoparorchiseurytremum. Res. Bull. Panjab. Univ. 44: 21-24.
- Dhingra, O. P., 1954c. Taxonomic values of chromosomes and cytoplasmic inclusions in a digenetic trematode, Phyllodistomum spatula. Res. Bull. E. Punjab Univ. 51: 101-109.
- Dhingra, O. P., 1955a. Spermatogenesis of a digenetic trematodeGastrothylaxcruminifer. Res. Bull. Panjab Univ. 65: 11-17.
- Dhingra, O. P., 1955b. Spermatogenesis of a digenetic trematodeCotylophoronelongatum. Res. Bull. Panjab Univ. 64: 1-10.
- Dhingra, O. P., 1955c. Gametogenesis and fertilization and cleavage in Asymphlodora sp. Res. Bull. E.Punjab Univ. 66: 19-29.
- Dvorak, J.,Vanacova, S.,Hampl, V.,Flegr, J. and Horak, P., 2002. Comparison of European Trichobilharzia species based on ITS1 and ITS2 sequences. Parasitology 124: 307-313.
- Fayaz, A. and Chishti, M. Z., 2000. Fish Trematode Parasites of Kashmir. Part II. Genus Clinostomum Leidy, 1856 (Digenea: Clinostomatidae). Oriental Sci. 5: 13-22.
- Frydrychova, R. and Marec, F., 2002. Repeated losses of TTAGG telomere repeats in evolution of beetles (Coleoptera). Genetic. 115: 179-187.
- Galazzo, D. E.,Dayanandan, S.,Marcogliese, D. J. and McLaughlin, J. D., 2002. Molecular systematics of some North American species of Diplostomum(Digenea) based on rDNA sequence data and comparisons with European congeners. Can. J. Zool. 80: 2207-2217.
- Graczyk, T., 1991. Variability of metacercariae of Diplostomumspathaceum(Rudolphi, 1819). (Trematoda,Diplostomidae). ActaParasitol. 36: 135-139.
- Gresson, R. A. R., 1964. Oogenesis in the hermaphroditic digenea (Trematoda). Parasitology, 54: 409-421.
- Gressоn, R. A. R. 1958. The gametogenesis of the digenetic trematode, Sphaerostomabramae (Muller) Luhe. Parasitol. 48: 293-302.
- Grossman, A. I. and Cain, G. D., 1981. Karyotypes and chromosome morphologies of Megalodiscustemperatus and Philophthalmusgralli. Journal of Helminthology, 55: 71-78.
- Guilford, H. G. 1955. Gametogenesis in Heronimuschelydrae. Trans. Amer. Mic. Soc. 74: 182-190.
- Guilford, H. G., 1961. Gametogenesis, egg-capsule formation, and early miracidial development in the digenetic trematodeHalipeguseccentricus Thomas. J. Parasitol. 47: 757-764.
- Gustinelli, A.,Caffara, M., Florio, D.,Otachi, E. O.,Wathuta, E. U. andFioravanti, M. L., 2010. First description of the adult stage of ClinostomumcutaneumPaperna, 1964 (Digenea: Clinostomidae) from grey herons Ardeacinerea L. and a redescription of the metacercaria from the Nile tilapia Oreochromisniloticusniloticus (L.) in Kenya. Syst. Parasitol. 76: 39-51.
- Hershkovitz, M. A. and Lewis, L. A., 1996. Deep-level diagnostic value of the rDNA-ITS region. Molecular Biology and Evolution 13: 1276-1295.
- Hillis, D. M. and Dixon, M. T., 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Quarterly Review of Biology66: 411-426.
- Jha, A. G., 1975. Cytogenetics, Evolution and systematic of Digenea. (Trematoda: Platyhelminthes).Egyptian journal of Genetics and Cytology 4: 201-233.
- Jones, A. W., 1956. The chromosomes of a species of HalipegusLooss, 1899 (Digenea: Hemiuridae). J. Tenn. Acad. Sci. 31: 186-187.
- Jousson, O. and Bartoli, P., 2002. Species diversity among the genus Monorchis (Digenea: Monorchiidae) parasitic in marine teleosts: Molecular, morphological and morphometrical studies with a description of Monorchisblennii n. sp. Parasitology Research 88: 230-241.
- Kane, R. A. and Rollinson, D., 1994. Repetitive sequences in the ribosomal DNA internal transcribed spacer of Schistosomahaematobium, Schistosomaintercalatum and Schistosomamattheei. Molecular and Biochemical Parasitology 63: 153-156.
- Kane, R. A.,Ridgers, I. L., Johnston, D. A. and Rollinson, D., 1996. Repetitive sequences within the first internal transcribed spacer of ribosomal DNA in schistosomes contain a Chi-like site. Molecular and BiochemicalParasitology 75: 265-269.
- Kanev, I.,Radev, V. and Fried, B., 2002. Family ClinostomidaeLuhe, 1901. In: D. I. Gibson, A. Jones, & R. A. Bray (Eds.), Keys to the Trematoda (Vol. 1). Wallingford, UK: CAB International and the Natural History Museum, pp: 113-120.
- Kaw, B. L., 1950. Studies in Helminthology: Helminth parasites of Kashmir. Part-I. Trematodes. Indian J. Helminthol. 2: 67-126.
- Levan, A. K.,Fredga, K. and Sandberg, A. A., 1964. Nomenclature for centromere position on chromosomes. Hereditas 52: 201-220.
- Luton, K., Walker, D. and Blair, D., 1992. Comparisons of ribosomal internal transcribed spacers from two congeneric species of flukes (Trematoda: Digenea). Mol. Biochem. Parasitol. 56: 323-328.
- Markell, E. K., 1943. Gametogenesis and egg-shell formation in ProbolitremacalifornienseStunkard, 1935 (Trematoda: Gorgoderidae). Trans. Am. Microsc. Soc. 62: 27-56.
- Matthews, D. and Cribb, T. H., 1998. Digenetic trematodes of the genus Clinostomum Leidy, 1856 (Digenea: Clinostomidae) from birds of Queensland, Australia, including C. wilsoni n. sp. from Egrettaintermedia. SystematicParasitology 39: 199-208.
- Morgan, J. A. T. and Blair, D., 1995. Nuclear rDNA ITS sequence variation in the trematode genus Echinostoma: an aid to establishing relationships within the 37-collar-spine group. Parasitology 111: 609-615.
- Mutafova, T. 1994. Karyological studies on some species of the families Echinostomatidae and Plagiorchiidae and aspects of chromosome evolution in trematodes. Systematic Parasitology, 28: 229-238.