Research Article - Journal of Clinical Pathology and Laboratory Medicine (2017) Journal of Clinical Pathology and Laboratory Medicine (Special Issue 1-2017)
Molecular characterization of plasmid mediated genes among S. aureus strains isolated from clinical and non-clinical sources, Ile-Ife, Nigeria.
- *Corresponding Author:
- Joseph Omololu-Aso
PhD Department of Microbiology, Obafemi Awolowo University, Ile-Ife, Osun Nigeria.
Tel: +2348133228723
E-mail: omololu-aso@oauife.edu.ng
Accepted date: April 25, 2017
Citation: Omololu-Aso J, Oluduro AO. Molecular characterization of plasmid mediated genes among S. aureus strains isolated from clinical and non-clinical sources, Ile-Ife, Nigeria. J Clin Path Lab Med. 2017;1(1):21-27.
Abstract
The study isolated and characterized clinical and non-clinical S. aureus strains in Ile-Ife, Nigeria using phenotypic and molecular methods. Eight hundred and fifty samples of different cultures were taken from clinical and nonclinical sources. The clinical sources were the routine specimens of wound swabs, urine, stool, blood and sputum from the Department of Microbiology and Parasitology laboratory of the Obafemi Awolowo University Teaching Hospitals Complex (OAUTHC) Ile-Ife. The non-clinical samples were obtained from the nasal cavity of apparently healthy food handlers at restaurants in Obafemi Awolowo University campus and food vendors in Ile-Ife central market. Samples were cultured on mannitol salt agar and incubated at 37°C for 24-48 hours. S. aureus were isolated and identified based on mannitol fermentation, Gram's reaction, positive results for catalase, coagulase and DNAse tests. Susceptibility of the isolates to nine different antibiotics was tested using the disk diffusion technique. Molecular detection of plasmid, mec A, nuc genes was carried out on representative isolates using polymerase chain reaction (PCR). The data generated were subjected to statistical analysis using T-Test. Of the 50 representative clinical and non-clinical isolates, 44% contained plasmid DNA with molecular weight ranging from 562 to 23,490 kb, while 18% had mec A and all selected representative isolates had the nuc gene. Only two (4%) out of the 50 S. aureus isolates investigated, were of PVL virulent strains attributed to urine sourced from both Pelvic inflammatory disease and wound of infected surgical implant individuals. These observations underscore the importance of confirming phenotypic identification of S. aureus by molecular techniques.
Keywords
S. aureus, Plasmid DNA, Molecular techniques, Molecular detection.
Introduction
Molecular methods are an improvement over conventional phenotypic methods for testing microbial relatedness in many ways. Currently, the most practical and useful application is in identifying outbreak of infections, determining modes and sources of acquisition of a pathogen, recognition of a particularly virulent strain of microorganism, identifying relapse of infection due to a single strain episodes of infections and effective preventive and therapeutic measures [1]. Phenotypic methods have occasionally been useful in most places describing the epidemiology of infectious diseases. DNA-based typing methods have eliminated the possible limitation and this is applicable for epidemiological studies [1]. A number of viable investigations indicated that S. aureus is the main etiological agent of many infections in Nigeria [2-7]. However, many studies, identification and antibiotic susceptibility testing of S. aureus isolates have been based on phenotypic methods and few data exist on the characterization of S. aureus using molecular methods [8-10].
The presence of the toxin Panton-Valetine (PVL) genes among S. aureus were first detected by PCR (Lina et al. 1999). Multilocus sequence typing (MLST), which was developed by using Neisseria meningitides as the model species [11], has been successfully adapted to S. aureus [12]. However, MLST is not suitable for routine surveillance of MRSA because of the high cost and the necessity of access to a high-throughput DNA sequencing facility.
Data on clinical identity, diversity, surveillance and new approaches in the molecular epidemiology of this pathogen in Nigeria are ongoing. This study was conducted for investigation on molecular characterization of virulent mediated genes among clinical and non-clinical isolate of S. aureus obtained from Ile- Ife, Nigeria.
Materials and Methods
Source of bacterial isolates
S. aureus isolates were recovered from both clinical and nonclinical specimens. The clinical sources were from the routine specimens of wound swabs, urine, stool, and sputum samples of the patients submitted to the Microbiology laboratory of the Obafemi Awolowo University Teaching Hospital Complex (OAUTHC), (Urban Centre), Ile-Ife, Nigeria.
The non-clinical isolates were recovered as nasal swabs from food handlers at the Obafemi Awolowo University (OAU) campus restaurants, marketers at the Ile-Ife Central Market and also from fomites within the hospital, which comprised of doctors stethoscopes and cell phones from the community dwellers and the Health care workers.
Collection of samples
Samples were collected between October, 2011 and November, 2012. Eight hundred and fifty (850) swab samples from clinical and non-clinical sources at Obafemi Awolowo University Teaching Hospital Complex, Obafemi Awolowo University campus community and Ile-Ife environs were obtained. Sterile cotton-tipped applicators (Sterilin, England) appropriately moistened with sterile distilled water were used for swabbing sample surfaces. Samples from both hospital (hospitalized patients who were on different types of antibiotic treatment) and the community of different sexes, age ranges and of different diagnostic infections histories ranging from diabetic ulcers, cancers (breast cancer, prostrate carcinoma) obstructive uropathy, (e.g. benign prostrate hypertrophy, septicaemia, urinary tract infection, burnt injuries, gastroenteritis, pelvic inflammatory diseases, sexually transmitted infections, pneumonia and other clinical diagnostic cases were considered. The non-clinical samples were obtained from the nostrils of apparently healthy community food handlers, cell phones and stethoscopes. The swabs were taken to the laboratory immediately for bacteriological analysis.
Microbiological analysis
Isolation of S. aureus was done by standard procedure in which the samples were inoculated on freshly prepared mannitol salt agar plates (Oxoid, Basingstoke, Hampshire, England) and incubated at 37°C for 24h. Golden yellow colonies on Mannitol Salt agar (MSA) after the incubation period were taken presumptively for S. aureus.
Phenotypic and biochemical identification of the isolates
Isolates were Gram stained as described by Olutiola et al. (1991). Biochemical identification of the isolates was carried out using standard methods. The following biochemical tests were carried out on the isolates namely catalase, tube coagulase test, and the DNase test using DNase agar based (Oxoid Ltd., Basingstoke, Hampshire, England). The confirmed isolates were stored as stock culture on nutrient agar slants and kept at about 4°C until further use.
Antibiotic susceptibility test
The susceptibility of the isolates to 8 selected antibiotics was carried out using the disc agar diffusion method according to National Committee for Clinical Laboratory Standards (Clinical Laboratory Standard Institute guidelines [9]. Freshly prepared Mueller-Hinton Agar (MHA) plates were flooded with 2 ml suspension of the test organisms in nutrient broth (106 cfu/ml). S. aureus ATCC 25923 was used as the control strain in every test run. The excess inoculums were drained up and culture plates were then left to stand for a few minutes. The antibiotics (Mast Diagnostic UK) employed included the followings: penicillin (10 μg), oxacillin (1 μg), cefoxitin (30 μg), gentamicin (10 μg), erythromycin (15 μg), clindamycin (2 μg), tetracycline (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg) and vancomycin (30 μg). Discs of the respective antibiotics were aseptically placed on the surface of the pre-inoculated agar plates using sterile forceps. Thorough contact of the discs with the agar was ensured by pressing the discs firmly but carefully on the plates with sterile forceps. There after culture plates were allowed to stand for 1hour to allow the antibiotics to diffuse into the agar medium and subsequently incubated at 37°C for 24 hours.
Plasmid isolation
Plasmid DNA of 50 randomly selected multiple antibiotic resistant S. aureus isolates was isolated as described by Birnboim and Doly, modified by use of lysostaphin for lysing the cell wall. Each S. aureus strain was inoculated into 3 ml trypton soy broth and incubated overnight on a roller drum at 37°C. About 1.5 ml of each overnight broth culture was transferred into Eppendorf tubes and centrifuged for one minute at 15000 rpm at room temperature. The supernatant was discarded and 2 μl of lysotaphin solution (1.0 μg/ml in distilled water) was added to the pellet. Tubes were capped, vortexed and placed on ice for 30 minutes. A 200 μl of alkaline detergent solution (0.2 N NaOH; 1% SDS) was added and the tubes inverted several times and then kept in the water bath for five minutes. Thereafter, 150 μl of 3M Sodium acetate (pH 4.8) was added and tubes were inverted several times to mix and then kept on ice for at least 10 minutes. The tubes were centrifuged at room temperature at 15,000 rpm for five minutes and the supernatant transferred into new Eppendorf tubes. One millilitre of 95% ice cold ethanol was added to the tubes, which were then kept at -20°C for five minutes. Thereafter, they were centrifuged at 15,000 rpm for three minutes; supernatant discarded and the sediment resuspended in 50 μl of Tris/ETDA (10 mM Tris HCl and 1 mM ETDA. pH 8.0). Thirty three microlitres of the contents were then loaded into wells of 1.0% agarose gels containing ethidium bromide. A 1.0 kb DNA ladder (MBI, Fermentas, Vilnius, Lithuania) was run side by side with test isolates as a molecular size marker. Electrophoresis was carried out in Tris acetate ETDA buffer containing ethidium bromide (20 ml of 50 × TAE and 6.0 μl of 10 μg/ml ethidium bromide per litre) at 30 mA (90 V) for four hours. Plasmid DNA was viewed with a UV transilluminator and photographs taken using a Leicaflex SL-camera. Films were exposed for 90 seconds and later developed. Plasmid sizes were estimated from a standard curve prepared from the molecular sizes of the 1.0 kb DNA ladder against their migration distance.
Molecular detection of the nuc and mec A genes by PCR
Phenotypically identified S. aureus isolates (only selected strains not all isolates) were confirmed as S. aureus by the detection of the nuc gene using the polymerase chain reaction (PCR). In addition, the presence of the mec A gene was determined to confirm the isolate as MRSA. Reference Primers (nuc-1) 5’AGTTCTGCAGTACCGGATTTGG-3’ (nuc-2) 5’-AAAATCGATGGTTGGC -3’ and (mec A1) 5’- CTC AGG TAC TGC TAT CCA CC; (mec-A2) 5’-CTC TTG GTA TAT CTT CAC C -3’ which amplified 280 bp and 449 bp segments of the nuc and mec A genes respectively were employed. Each PCR assay was made up of the following: 25 μl of mastermix (Sigma) containing 1.5 units of Taq DNA polymerase, 10 mM Tris-HCl, 50 mM KCl, 1.5 Mm MgCl2, 0.001% w/v gelatin and 0.2 mM dNTPs, 1 μl (20 pmol) of the forward and reverse primers and 5 μl of template DNA. The thermocycler was programmed with the following parameters: pre-denaturation at 95°C for 5 minutes, 30 cycles of denaturation at 95°C for 30 seconds, extension at 72°C for 1 minute and final extension at 72°C for 5 min.
The PCR conditions were utilized for the detection of the nuc A, and mec A genes. PCR products were detected by gel electrophoresis using 1.5% (w/v) agarose. The agarose gel was run in 1XTBE 0.089M Tris 0.089 M Boric acid, 0.002 M EDTA disodium buffer (pH 8.3) for 2 hrs at 80 V. Thereafter, the gels were stained with ethidium bromide and visualized under UV light.
Separation of amplified products by agarose gel electrophoresis
Three grams of agarose was weighed and added to a conical flask containing 250 ml of 1x TAE buffer. The contents were melted by heating in an oven and the solution was stirred for proper mixing and complete dissolution of agarose. The agarose gel solution was cooled to about 40°C to 45°C and 2-3 drops of Ethidium bromide (0.5 μg/ml) was added. Agarose gel was poured into the casting platform after inserting the comb in tray. While pouring, sufficient care was taken to prevent the formation of air bubbles. The gel was allowed to solidify and the comb was removed after placing the solidified gel into the electrophoretic apparatus containing sufficient buffer (1x TAE), so as to cover the wells completely. About 2.5 μl of loading dye was added to each tube containing amplified DNA. The amplified products (20 μl) were carefully loaded into the sample wells. Electrophoresis was carried out at 60 volts until the tracking dye migrated to the negative end of the gel. Gel was viewed under UV trans-illuminator for DNA bands and the photographed for documentation.
Scoring of amplified fragments
The amplified profiles for the primer were compared with each other and bands of DNA fragment were scored as ‘l’ for presence and ‘0’ for absence, generating ‘0’ and ‘1’matrix. Percent polymorphism was calculated by using the formula:
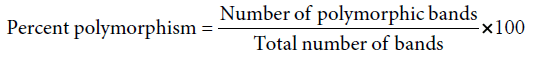
Statistical analysis of data
Statistical analysis methods to determine frequency distribution, mean, harmonic mean, standard deviation, analysis of variance using T-test correlation was employed
Results
Staphylococccus aureus isolates
A total of 405 S. aureus isolates were obtained from the 721 staphylococci recovered from 850 samples collected from clinical and non-clinical sources. The clinical isolates comprised of 230 (57%) and 175 (43%), non-clinical. clinical isolates were sourced as follows; wounds (58); stools (47); urine (58); sputum (37) and blood (30), The highest rate of isolation of S. aureus isolates from clinical sources was from wounds (14.3%) and stools samples (14.3%), while cell phones 15% and food handlers (15%) constituted the highest among non-clinical S. aureus isolates. Overall, the prevalence of S. aureus isolates recovered from clinical sources are of statistical different (T= 0.141).
Plasmid Profile of S. aureus isolates
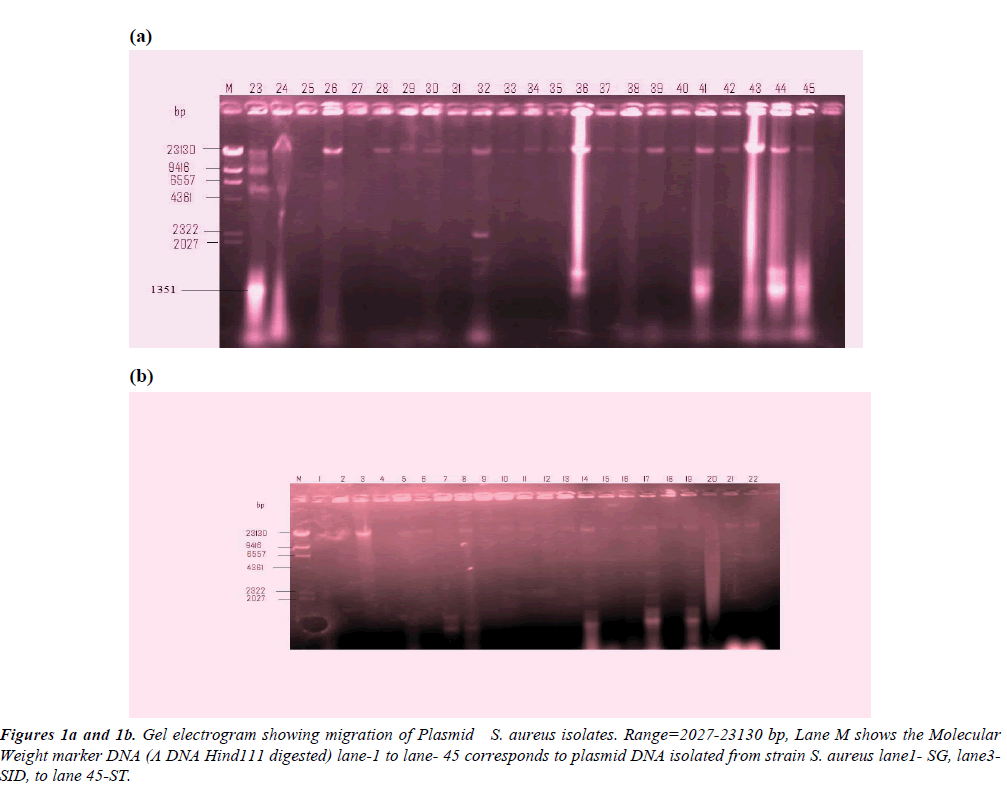
Table 1 shows the fragment lengths plasmids polymorpho-types of the 50 randomly selected MAR isolates. The gel electrogram showing the plasmid DNA bands and their molecular weight is depicted in Figures 1a and 1b. Molecular weight marker DNA (Λ DNA Hind111 digested) was employed. The molecular weight of the plasmid isolates ranged between 562 kb to 23,490 kb. Among the clinical isolates, 24% carried plasmid gene in which S. aureus isolates from blood sepsis, chronic renal failure, and wound post-operative infected individual showed multiples of 4, 4, and 3 serrative bands indicating plasmids of varied molecular weight (23,490 kb; 9.416 kb; 6.55 7kb; 1,351) 23,490 kb, 2,322 kb, 21,027 kb, 562 kb) and (23,490 kb, 2,322 kb. 1’351 b) respectively. However, wound infections were the most frequent isolates harboring plasmid among the clinical sources (Figure 2). Meanwhile, 20% of the isolates from the non-clinical carried 20% of total plasmids trait in which stethoscopes appeared the most frequent harbored of plasmid DNA. Double serrative bands were noticed among stethoscopes and cellphones S. aureus isolates, with varied molecular weights (9,416 kb; 4,361 kb) and 23,490 kb.562 kb) respectively are shown in Table 1.
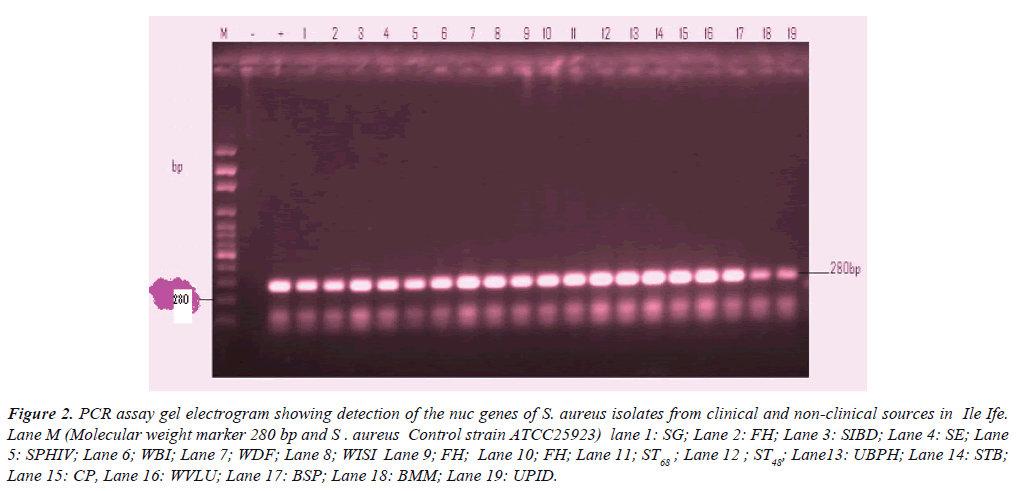
Figure 2:PCR assay gel electrogram showing detection of the nuc genes of S. aureus isolates from clinical and non-clinical sources in Ile Ife. Lane M (Molecular weight marker 280 bp and S . aureus Control strain ATCC25923) lane 1: SG; Lane 2: FH; Lane 3: SIBD; Lane 4: SE; Lane 5: SPHIV; Lane 6; WBI; Lane 7; WDF; Lane 8; WISI Lane 9; FH; Lane 10; FH; Lane 11; ST68 ; Lane 12 ; ST48; Lane13: UBPH; Lane 14: STB; Lane 15: CP, Lane 16: WVLU; Lane 17: BSP; Lane 18: BMM; Lane 19: UPID.
| TYPES: A B C D E F G H I J | |||||||
|---|---|---|---|---|---|---|---|
| Lane | Single | Double | Triple (3) | Tetra(4) | |||
| Food handlers | 3 | 28, 29, 30 | J(3)100% | __ | _ | _ | |
| Stethoscope | 4 | 8, 45 | J(1) 25% | GI(1),25%CJ(2), 50% | _ | _ | |
| Cell phones | 3 | 3, 14, 19 | J(1)33% | AJ(1), 33% BJ(1),12.5% |
_ | _ | |
| Wound | 8 | 17, 21, 22, 29, 34, 35, 43, 44 | J(5) 62.5% | BJ(1),12.5%CJ(1),12.5% | CFG (1) 12.5% |
_ | |
| Blood | 2 | 23, 26 | J(1) 50% | __ | _ | CHIJ(1) 50% |
|
| Urine | 2 | 32. 36 | _ | DJ(1) 50% |
_ | AEFG(1) 50% |
|
| Total | 11 | 8 | 1 | 2 | |||
|
|||||||
Table 1: Fragment lengths plasmids polymorpho-types.
Molecular detection of nuc genes
Table 2 shows the detection of nuc gene in clinical and nonclinical isolates previously identified phenotypically as S. aureus in relation to the patients’ age, sex and case diagnosis. All the 50 selected representative isolates had the nuc gene.
| Isolates codes | PatientsAge | Sex | Sources | Case diagnosis | electrophoretic lane designation |
|---|---|---|---|---|---|
| SG | 60 | F | Stool | Gastroenteritis | 1 |
| FH | 21-30 | M | Nostril | - | 2 |
| SIBD | 22 | M | Stool | Inflammatory bowel disease | 3 |
| SE | 40 | F | Stool | Enterocolitis | 4 |
| SPHIV | 35 | M | Sputum | HIV | 5 |
| BI | 20 | M | Burnt injury | 6 | |
| DF | 53 | F | Diabetic foot | 7 | |
| ISI | 45 | F | Infected surgical implant | 8 | |
| FH | 51-60 | - | - | 9 | |
| FH | 31-40 | - | - | 10 | |
| ST68 | - | - | - | 11 | |
| ST48 | - | - | - | 12 | |
| UBPH | 64 | F | Urine | Benign prostatic hypertrophy | 13 |
| STB | 46 | M | Sputum | Tubercullosis | 14 |
| CP | - | - | Cellphone | - | 15 |
| WVLU | 60 | M | Wound | Venousleg ulcer | 16 |
| BSP | 18 | F | Blood | Septicaemia | 17 |
| BMM | 45 | F | Blood | Multiple myeloma | 18 |
Table 2: Molecular detection of nuc gene from clinical and non-clinical isolates of S. aureus.
Molecular detection of mec A genes
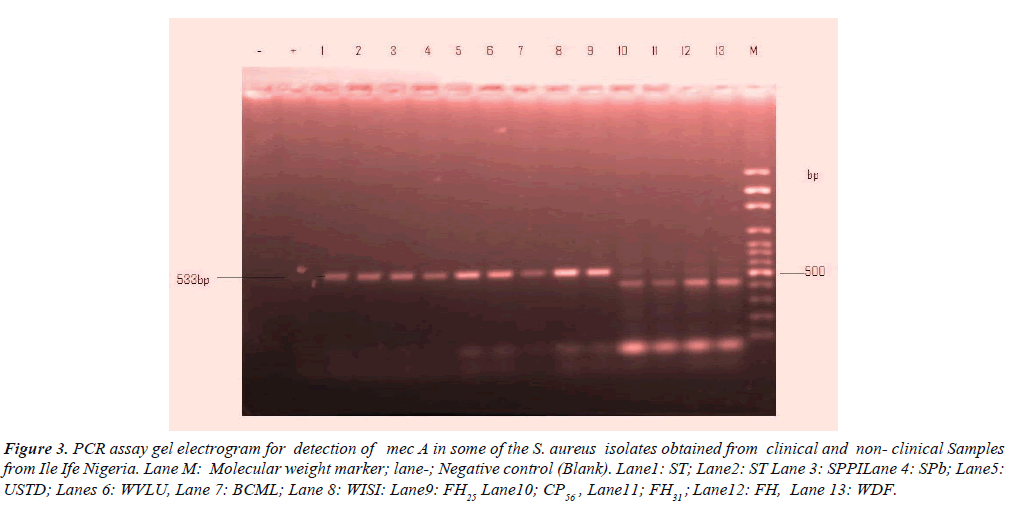
Table 3, shows detection of mec A gene in the clinical and nonclinical S. aureus isolates. A total of nine 9 (18%) among 50 representative isolates contained mec A gene of which wound, sputum, stethoscope predominated each by 2(4.0%) followed by urine, stethoscopes and food handlers 1(2.0%) respectively. However, female individuals overwhelmingly constituted 5(10.0%) and male 2(4.0%) of mec A respectively. The distinct image clearly visible in Figure 3 is the electrophoresis gel banding patterns.
Figure 3: PCR assay gel electrogram for detection of mec A in some of the S. aureus isolates obtained from clinical and non- clinical Samples from Ile Ife Nigeria. Lane M: Molecular weight marker; lane-; Negative control (Blank). Lane1: ST; Lane2: ST Lane 3: SPPILane 4: SPb; Lane5:USTD; Lanes 6: WVLU, Lane 7: BCML; Lane 8: WISI: Lane9: FH25 Lane10; CP56 , Lane11; FH31 ; Lane12: FH, Lane 13: WDF.
| Isolates codes | Patients Age | Sex | Sources | Case diagnosis | Electrophoretic lane designation |
|---|---|---|---|---|---|
| ST | - | - | Stethoscope | - | 2 |
| SPPI | 50 | M | Sputum | Pulmonary inflammation | 3 |
| SPB | 60 | F | Sputum | Chronicbronchitis | 4 |
| USTD | 35 | F | Urine | Sexually transmitted disease | 5 |
| WVLU | 19 | M | Wound | Venous leg ulcer | 6 |
| BCML | 50 | F | Blood | Chronic myeloma | 7 |
| WISI | 27 | F | Wound | Postoperative. | 8 |
| FH25 | 51-60 | F | Nostril | - | 9 |
| L |
Table 3: Molecular detection of mec A gene from clinical and nonclinical isolates of S. aureus
Discussion
In the present study, 22(44%) of the clinical and non-clinical S. aureus isolates screened for the presence of plasmid harboured plasmid mediating factors of molecular weight ranging from 562 kb to 23,490 kb in which 24% majorly from wound isolates were obtained from the representative clinical sources and 20% originated from non-clinical sources among which stethoscope the associated fomites sourced isolates were frequently implicated. This finding is in consonance with the report of [13] and [14] which stated that resistance to high levels of antibiotics has been attributed in most instances to the presence of plasmids. This was also corroborated by other workers [13,15-16]. Plasmids were not detected in 28 of the resistant strains indicating that their resistance was probably chromosomally mediated. Different patterns of antibiotic resistance and plasmid profiles among strain of S. aureus have been reported [17]. Staphylococcal antibiotic resistance has been associated with resistance plasmids that have the ability to mediate the production of drug inactivating enzymes such as β-lactamases (Adeleke and Asani, 2009) and other functions. The emergence of resistance plasmids in this study could be due to overzealous desire to treat every infection diagnosed or undiagnosed and over the counter availability of antibiotics [18]. Detection of S. aureus in the community food handlers’ nasal cavity in 10 (20%) of the samples screened was lower to that reported by [19] and Daini and Akano, (2009). All the 50 selected representative isolates were positive for nuc gene. The detection of the nuc gene is the gold standard for the identification of S. aureus. Mis-identification of bacterial pathogens has dire consequences on the patients. This includes increased medical cost and prolonged stay among other attendant consequences. This observation underscores the importance of confirming phenotypic identification of S. aureus by molecular techniques. Among 50 representative isolates in this study, mec A gene was detected in 9 which constituted 18% of the MRSA isolates. This was lower compared to the study conducted by the Association for Professionals in Infection and Epidemiology [20] on the prevalence of MRSA in the US. Health care facilities. They showed that patients infected with MRSA account for as many as 50% to 70% of the S. aureus infections acquired in health care facilities (APIE, 2007). Although multiple methods of detection of methicillin resistance have been developed, identification of the mec A gene is the most reliable reference method for detecting MRSA isolates The MRSA prevalence rate of 18% observed in this study among the 50 screened representative isolates from clinical sources is further in support of the report of [21-24] in which associated MRSA is implicated with high morbidity and mortality potential for intra and inter- hospital dissemination and the spread of epidemic and nosocomial infections worldwide. Molecular detection of mec A has been described as ‘gold standard’ for confirming MRSA, hence Microbiological Laboratory should be encouraged to develop capacity for the routine identification and confirmation of MRSA based on the molecular detection of the mec A gene.
References
- Pfaller MA, Hewald LA. Diagnosis and management of infectious diseases: Molecular methods for the new millennium. Clin Lab News. 2000;26:10-13.
- Ako-Nai K, Adejuyigbe J, Ajayi V, et al. The bacteriology of neonatal septicaemia in Ile-Ife, Nigeria. J trop pediatr. 1999;45(3):146-51.
- Anah MU, Udo JJ, Ochigbo SO, et al. Neonatal septicaemia in Calabar, Nigeria. Tropical doctor. 2008;38(2):126-8.
- Odetoyin WB, Aboderin AO, Ikem RT, et al. Asymptomatic bacteriuria in patients with diabetes mellitus in Ile-Ife, South-West, Nigeria. East Afr med j. 2008;85(1):18.
- Adeleke SI, Asani MO. Urinary tract infection in children with nephrotic syndrome in Kano, Nigeria. Ann Afri med. 2009;8(1):38.
- Bekibele CO, Kehinde AO, Ajayi BG. Upper lid skin bacterial count of surgical eye patients in Ibadan, Nigeria. Afr j med med sci. 2008;37(3):273-7.
- Onipede AO, Onayade AA, Elusiyan JB, et al. Invasive bacteria isolates from children with severe infections in a Nigerian hospital. J Infec Dev Countr. 2009;3(06):429-36.
- Adesida S, Boelens H, Babajide B, et al. Major epidemic clones of Staphylococcus aureus in Nigeria. Microb Drug Resist. 2005 Jun 1;11(2):115-21.
- Shittu AO, Lin J. Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in KwaZulu-Natal province, South Africa. BMC Infec dis. 2006;6(1):125.
- Okon KO, Basset P, Uba A, et al. Cooccurrence of predominant Panton-Valentine leukocidin-positive sequence type (ST) 152 and multidrug-resistant ST 241 Staphylococcus aureus clones in Nigerian hospitals. J clin microbiol. 2009;47(9):3000-3.
- Maiden MC, Bygraves JA, Feil E, et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140-5.
- Enright MC, Robinson DA, Randle G, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci. 2002 May 28;99(11):7687-92.
- Diep BA, Gill SR, Chang RF, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. The Lancet. 2006;367(9512):731-9.
- Han K, Jang SS, Choo E, et al. Prevalence, genetic diversity, and antibiotic resistance patterns of Campylobacter jejuni from retail raw chickens in Korea. Int J Food Microbiol. 2007;114(1):50-9.
- Bhakta M, Arora S, Bal M. Intraspecies transfer of a chloramphenicol-resistance plasmid of staphylococcal origin. Ind J Med Res. 2003;117:146.
- Daini OA, Akano SA. Plasmid-mediated antibiotic resistance in Staphylococcus aureus from patients and non patients. Sci Res Essays. 2009;4(4):346-50.
- Akinyemi KO, Alabi SA, Taiwo MA, et al. Antimicrobial susceptibility pattern and plasmid profiles of pathogenic bacteria isolated from subjects with urinary tract infections in Lagos, Nigeria. Niger. Qtr J Hosp Med. 1997;1:7-11.
- Diani OA, Ogbolu DO, Ogunledun A. Plasmid determined resistance to quinolones in clinical isolates of Gram-negative bacilli. Afr j med med sci. 2006;35(4):437-41.
- Hidron AI, Kourbatova EV, Halvosa JS, et al. Risk factors for colonization with methicillin-resistant Staphylococcus aureus (MRSA) in patients admitted to an urban hospital: Emergence of community-associated MRSA nasal carriage. Clin infect dis. 2005;41(2):159-66.
- Association of Professionals in Infection and Epidemiology (APIE), (2007). National prevalence study of methicillin-resistant Staphylococcus aureus (MRSA) in U.S. healthcare facilities (Executive summary), 34th APIC Conference, 2007. San Jose California.
- Vaez H, Tabaraei A, Moradi A, et al. Evaluation of methicillin resistance Staphylococcus aureus isolated from patients in Golestan province-north of Iran. Afr J microbiol res. 2011;5(4):432-6.
- Wikler MA. Performance standards for antimicrobial susceptibility testing: Eighteenth informational supplement. Clinical and Laboratory Standards Institute (CLSI); 2008.
- Panton PN, Valentine FC. Staphylococcal toxin. The Lancet. 1932;219(5662):506-8.
- Strommenger B, Braulke C, Heuck D, et al. Spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J Clin Microbiol. 2008;46(2):574-81