Research Article - Asian Journal of Biomedical and Pharmaceutical Sciences (2020) Volume 10, Issue 70
First phenotypic and molecular characterization of clinical isolates of drug resistant Providencia rettgeri in Imo state, South-Eastern Nigeria.
Malachy C Ugwu1*, Lynda U Ndunagu1, Chika P Ejikwugwu1,2, Charles O Esimone1
1Department of Pharmaceutical Microbiology and Biotechnology, Nnamdi Azikiwe University, Awka, Nigeria
2Department of Applied Microbiology, Ebonyi State University Abakiliki, Ebonyi State, Nigeria
- Corresponding Author:
- Malachy C Ugwu
Department of Pharmaceutical Microbiology and Biotechnology
Nnamdi Azikiwe University
Awka, Nigeria
E-mail: mc.ugwu@unizik.edu.ng
Accepted date: June 22, 2020
DOI: 10.35841/2249-622X.70.12507
Visit for more related articles at Asian Journal of Biomedical and Pharmaceutical SciencesAbstract
Background: Nosocomial outbreak of drug resistant Enterobacteriaceae including those mediated by Providencia species has become a serious public health challenge across the globe – owing to the
difficulty in selecting antibiotics for proper treatment especially in the face of resistance. The morbidity
and mortality rate of bacteremia due to Providencia species could be high, especially among the elderly
and those with pre-existing medical conditions. This study was aimed at determining the occurrence of
multidrug resistance phenotypes in clinical isolates of Providencia rettgeri using both phenotypic and
genotypic techniques.
Methods: Clinical isolates were identified to be isolates of P. rettgeri using standard microbiological and
biochemical techniques; and these were characterized with respect to antibiotic resistant patterns.
Overall, 143 clinical isolates of non-duplicate P. rettgeri isolates were bacteriologically recovered from
the clinical specimens which included blood/wound swab samples (n=108) and swabs from surgical
instruments and suction devices (n=35). Antimicrobial susceptibility test (AST) was done using the
modified Kirby-Bauer disk diffusion techniques as per the Clinical Laboratory Standard Institute
(CLSI) criteria. The isolates were tested phenotypically for ESBL production by the double disk synergy
test (DDST) method and the β-lactamase genes were identified using polymerase chain reaction. The
strains were sequenced with ABI3500XL analyzer with a 50 cm array, using POP7. Sequences data
generated were analyzed with Geneious version 9. 0. 5.
Results: The result of AST showed that Providencia rettgeri isolates were resistant to ceftazidime,
cefotaxime, ampicillin and tetracycline. Based on the DDST method, some of the Providencia rettgeri isolates were phenotypically confirmed to be ESBL positive (11%) and MBL positive (6%). The results
of the molecular analysis showed that the P. rettgeri isolates harbored SHV genes (n=6; 12. 5%) which
mediates ESBL production. However, none of the isolates harboured the CTX-M or TEM gene.
Conclusion: Our study shows that the P. rettgeri isolates analyzed were multidrug resistant, and they
also harbour some important phenotypes and/or genotypes that mediate the production of ESBL
(extended spectrum beta-lactamase). This is the first report of ESBL detection in clinical isolates of P. rettgeri in Imo state, southeast Nigeria.
Keywords
Providencia rettgeri, Enterobacteriaceae, Antibiotic resistance, ESBL, Nigeria.
Abbreviations
AMP: Ampicillin; AMC: Amoxicillin-Clavulanic Acid; CAZ: Ceftazidime; CTX: Cefotaxime; CIP: Ciprofloxacin; CRO: Ceftriaxone; CN: Gentamicin; OFX: ofloxacin; SXT: sulphamethoxazole/trimethoprim; TE: Tetracycline.
Introduction
The genus Providencia is a member of Enterobacteriaceae family and thus forms part of the normal gastrointestinal flora of humans and animals [1,2]. It is an aerobic Gram-negative bacillus and belongs to Proteeae bacteria [3]. It is made up of five species: Providencia alcalifaciens, Providencia rustigianii, Providencia stuartii, Providencia rettgeri Providencia rettgeri and Providencia heimbachae [4,5]. Among the Providencia species, Providencia rettgeri is the most common cause of infections in humans. P. rettgeri causes nosocomial infections in humans [5]. It is implicated as a causative agent in urinary tract infections, bacteraemia, gastroenteritis, skin and wound infections [1]. P. rettgeri has also been implicated in traveler’s diarrhea and more severe human infections like meningitis [1,6]. P. rettgeri is known to harbour various virulence and antibiotic resistance genes and thus treatment of P. rettgeri borne infections is challenging [1,2]. The most noted mechanism of resistance to antibiotics is commonly by the production, n of an enzyme that breaks down the antibiotic before it can affect the bacterium [1]. Monitoring the prevalence of antibiotic resistant Enterobacteria of clinical interest may contribute to assuaging the nefarious activities of these organisms, and thus help to guide therapy for those already infected. Thus, the detection of the underlying resistance mechanisms in these organisms is critical for better management of P. rettgeri borne infections. It is in view of these challenges that this study was designed to characterize and determine the antibiotic resistance profile of the clinical isolates of P. rettgeri in Owerri metropolis, Imo state, Nigeria.
Methods
Ethical consideration
The study protocol was performed according to the Helsinki declaration and approved by Imo state Ministry of health and the management of the Hospital.
Sample collection and processing
One hundred and forty three clinical samples (143) which included seventy (70) blood specimen, wound swabs (n=38) taken from patients admitted into these selectedhospitals within Owerri metropolis and thirty five (35) samples obtained from surgical instruments and suction devices were recruited for this study. All samples were aseptically inoculated in nutrient broth (Oxoid, UK) and incubated overnight at 37°C. After incubation bacterial growth was confirmed by the presence of visible bacterial growth as evidenced by turbidity. All turbid tubes were each sub-cultured onto freshly prepared plates of blood agar and MacConkey agar media. The culture plates (Oxoid, UK) for the selective isolation of P. rettgeri isolates and was incubated overnight at 37°C. The presence of P. rettgeri on the culture plates was determined qualitatively based on colonial morphology or characteristics, microscopy, and biochemical testing [7,8].
Antibiotic susceptibility testing
Antibiotic disk containing ciprofloxacin (5 μg), ofloxacin (5 μg), gentamicin (10 μg) cefotaxime (30 μg), ceftazidime (30 μg), cefoxitin (30 μg), ceftriazone (30 μg)vancomycin (30 μg) , oxacillin (l μg), cloxacillin (5 μg), amoxicillin-clavulanic acid (20/10 μg) and ampicillin (10 μg), imipenem (10 μg), meropenem (10 μg) and trimethoprim-sulfamethoxazole (1. 25/23. 75 μg) were aseptically placed on Mueller-Hinton (MH) agar plates previously inoculated with the test organism. The plates were incubated at 37°C for 18-24 hours and the Inhibition Zone Diameter (IZDs) produced were measured , recorded and interpreted followingthe guidelines of the Clinical Laboratory Standard [9].
Double disk synergy test (DDST) for detection of extended spectrum beta lactamase (ESBL)
The P. rettgeriisolates with diameter zone of inhibition of ≤ 22 mm for ceftazidine and ≤ 25 for ceftriaxone were further screened for ESBL production by DDST on Muller-Hinton (MH) agar (Oxoid, UK) plates as previously described [10,11]. ESBL production was confirmed when there is an increase of ≥ 5 mm in inhibition zone diameter for either of the cephalosporins (ceftazidime and cefotaxime) tested in combination with amoxycillin-clavulanic acid versus its zone when tested alone.
Evaluation of metallo-β-lactamase (MBL) production by the P. rettgeri isolates
MBL was detected phenotypically by subjecting the imipenem resistant isolates to combined disc test. An organism was considered to be MBL positive if there was an increase of ≥ 7 mm in the zone of inhibition around the imipenem+EDTA disc as compared to imipenem disc alone after incubation at 37°C for 18-24 hours as was previously described [12].
Multiplex PCR characterization of ESBL genes’
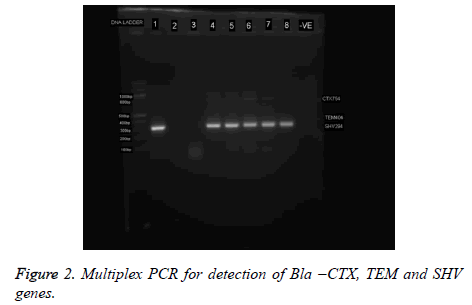
Table 1 shows the gene sequence of primers (forward and reverse) used for the amplification of Bla –CTX, TEM and SHV genes. An aliquot of 1 μl of template DNA was added to 19 μl of master mixture containing 13 μl of the following reagents: dNTP mixture, PCR buffer, Taq polymerase (250 μl), nuclease free water, and 0. 1 μl of each forward and reverse primers (6 μl). The prepared PCR tubes with master mixture were placed in the Bio-Rad I thermal cycler (UK). Amplification of the ESBL genes was carried out according to the following thermal cycling condition Initial denaturation (95°C for 5 minutes), followed by 40 cycles of denaturation (95°C for 30 seconds), annealing (51°C for 1 hour, 30 seconds) and extension (72°C for 30 seconds ), with a single final extension (at 72°C for 2 minutes). The PCR product (amplicon) was separated by gel electrophoresis and visualized using a 100 bp DNA molecular size marker.
| Primer Name | Sequence 5’–3’ | Size (bp) |
|---|---|---|
| 294 –F | CGC TCT TCT TAG GAA CTC TG | 294 |
| CTX –R | GGC TGG GTG AAG TAA GTG AC | |
| SHV –F | CGC CTG TGT ATT ATC TCC CT | 404 |
| SHV –R | CGA GTA GTC CAC CAG ATC CT | |
| TEM –F | CGA CGA GTG GGT TAC ATC GA | 754 |
| TEM -R | GCT CCT CCG ATC GTT GTC AG |
Table 1: Sequence of primers used.
PCR product cleaning and purification
The PCR products were cleaned using Exo/SAP. The Exo/SAP Master mix was prepared by adding 50. 0 μl Exonuclease I (NEB M0293) 20 U/ul and 200. 0 μl Shrimp Alkaline Phosphatase (NEB M0371) 1 U/ul in a 0. 6 ml microcentrifuge tube. Exo/SAP Mix of 2. 5 μl was then added to 10. 0 μl of PCR Mixture, mixed well and was incubated at 37°C for 30 minutes after which the reaction was stopped by heating at 95°C for 5 minutes. Purification was done with the ABI V3. 1 Big dye kit according to manufacturer’s instructions. The labeled products were then cleaned with the ZymoSeq clean-up kit. Sequencing Binding Buffer of 240 μl was added to 20 μl sequencing reaction which was then transferred to a Zymo- Spin ™ IB-96 plate mounted onto a collection plate. The mixture was centrifuge at 3, 000 xg for 2 minutes. Sequencing Wash Buffer of 300 μl was added to each well of the plate and centrifuge at 3, 000 xg for 5 minutes. A 15 μl of nuclease free water was directly added to the column matrix of the filter plate. The ZymoSpin™ IB-96 plate was placed on top of the supplied 96-Well PCR plate and mounted on the assembly onto the collection plate which was then centrifuged at 3, 000 xg for 2 minutes to elute the DNA.
Sequencing
The Ultra-pure DNA was sequenced with ABI3500XL sequence analyzer (Thermofisher, China) with a 50 cm array, using POP7 at Inqaba Biotechnical Industries Ltd (Hatfield, South Africa). The gene sequence data generated were analyzed with Geneious software version 9. 0. 5 Phylogenetic tree were constructed using neighbor joining.
Statistical analysis
The data were analyzed using student t-test with the aid of SPSS 22 software package and expressed as mean values ± (Standard error of the mean) of the three replicates of antibiotic susceptibility profile of the isolated bacteria against panel of antibiotics.
Results and Discussion
Providencia rettgeri has been linked with the nosocomial infections of the wounds, urinary tract burns and blood. The incidence rate of P. rettgeri isolates from clinical samples was 33. 5% (Table 2).
| S/N | Sample | No of samples examined | Number of isolates | Occurrence rate (%) |
|---|---|---|---|---|
| 1 | Blood | 70 | 3 | 4. 3 |
| 2 | Burn Surface Area (BSA) | 38 | 31 | 81. 6 |
| 3 | Surgical blades | 9 | 4 | 44. 4 |
| 4 | Sinks | 5 | 2 | 40 |
| 5 | Others (Suture, Lab coat, disposable gloves) | 21 | 8 | 25. 8 |
| Total | 143 | 48 |
Table 2: Detection rate of P. rettgeri isolates from clinical samples.
It has been stated that sources of Providencia are varied and each species of Providencia have a preferred habitat [5]. In our study, Burn Surface Area (BSA) accounted for more (64. 6%) of the isolation sites. This might explain why P. rettgeri has been reported to cause wound and burn infections.
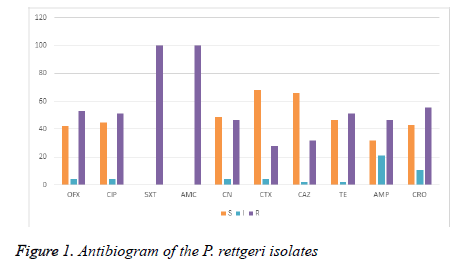
All the test isolates were resistant to sulphamethoxazoletrimethoprim (SXT) and amoxycillin-clavulanic acid (AMC) acid. Almost all the strain of Providencia rettgeri was resistant to ceftazidime, cefotaxime, ampicillin and tetracycline. Particularly, Providencia rettgeri isolates (Pr2, Pr7, Pr9, Pr16, Pr34, Pr41 and Pr46) were resistant to the 3rd generation cephalosporin (ceftazidime, cefotaxime, ceftriaxone). Providencia rettgeri has been previously reported to be often resistant to multiple antibiotics [5]. In a study conducted in Ile- Ife, Southwestern Nigeria, it was found that all Providencia species were sensitive to cephalosporins including ceftazidime, cefuroxime, and cefixime [13,14]. Strikingly, majority of the isolates in our study had MARI greater than 0. 2. In this study, we found a 74% resistance to third generation cephalosporin among the Providencia rettgeri isolates recovered from patients in Owerri , Imo state (Figure 1) . In the study location, during emergencies and as antibiotic coverage during surgery, cephalosporins especially ceftriaxoneare administered empirically. This practice may have over time exerted selective pressure, resulting in the emergence and dissemination of ESBL-positive bacteria in the hospital environment.
Extended-Spectrum β-Lactamases (ESBLs) and Metallo- Betalactamases (MBL) have been recognized as key causes of resistance in Gram-negative bacteria. Acquired resistance by the production of ESBL and MBL enzymes is a common reported mechanism among the Enterobacteriaceae [15-19]. Phenotypically, eleven (11%) of the isolates were ESBL positive while 6% were MBL positive (Table 3). Though, the coexistence of ESBL with other types of β-lactamases is no morea rare event, only one isolate co-harbored ESBL and MBL in this study (Table 3). A similar concomitant presence of 2 β- Lactamases in one strain has been reported by Ibadin et al. [20] in Southwestern Nigeria. The co-existence of ESBL and MBL enzymes in a bacterium will severely limit treatment options in treating infections caused by the strain [19-21]. It should be noted that these enzymes are hardly screened for, during routine laboratory susceptibility testing of clinical bacterial isolates in healthcare facilities in Nigeria. And since routine susceptibility studies carried out in hospital laboratories are deficient in detecting ESBL- or MBL-producing bacteria from clinical samples, proper ESBL- and/or MBL- detection protocol should be added to the routine susceptibility studies in hospital laboratories so that ESBL- and MBL-producing bacteria can be adequately detected, reported and contained (Figure 2).
| S/No. | Beta Lactamase Enzyme | No. positive (%) |
|---|---|---|
| 1 | ESBL | (5) 11% |
| 2 | MBL | (3) 6% |
| 3 | ESBL+MBL | (1) 2% |
Table 3: Frequency of ESBL and MBL Production From 47 P. rettgeri isolates From Clinical Specimens.
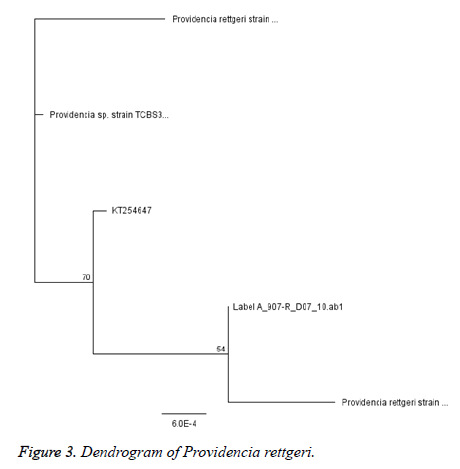
Six (12. 5%) of the P. rettgeri isolates harbored SHV genes while none of the isolates harbored CTX-M or TEM (Figure 3). It is possible to presume that bla SHV genes were responsible for the ESBL phenotype observed (Figure 2 and Table 3). A similar non detection of CTX-M ESBL among isolates of P. rettgeri was reported by Dropa et al. , [21]. The phylogenetic tree revealed that all isolates are P. rettgeri.
Conclusion
Our study shows that the P. rettgeri isolates analyzed were multidrug resistant, and they also harbour some important phenotypes and/or genotypes that mediate the production of ESBL (extended spectrum beta-lactamase). We reported that bla SHV was the most commonly detected antibiotic resistance genes. This is the first report of ESBL detection in clinical isolates of P. rettgeri in Imo state, southeast Nigeria.
Competing Interests
The authors declare that they have no competing interests.
Authorship and Contributions
Conceptualization of the study: COEsimone and MC Ugwu
Data generation: LUNdunagu
Writing, Review and Editing of the manuscript:MCUgwu and CP Ejikeugwu
References
- Tshisevhe VS, Lekalakala MR, Tshuma N, et al. Outbreak of carbapenem-resistant Providencia rettgeri in a tertiary hospital. S Afr Med J. 2017;107: 31-33.
- Zavascki AP, Carvalhaes CG, Da SGL. Outbreak of carbapenem-resistant Providencia stuartii in an intensive care unit. Infect Control HospEpidemiol. 2012;33:627-30.
- Olaitan AO, Diene SM, Assous MV, et al. Genomic plasticity of multidrug-resistant NDM-1 positive clinical isolate of Providencia rettgeri. Genome BiolEvol. 2016; 8:723-28.
- Abdallah M, Alhababi R, Alqudah N, et al. First report of carbapenem-resistant Providencia stuartii in Saudi Arabia . New Microb New Infect. 2018; 26: 107–9
- Sharma D, Sharma P, Soni P. First case report of Providencia rettgeri neonatal sepsis. BMC Res Notes. 2017; 10:536: 1-3
- Barrios H, Garza-Ramos U, Reyna-Flores F, et al. Isolation of carbapenem-resistant NDM-1-positive Providencia rettgeriin Mexico. J AntimicrobChemother. 2013; 68:1934-36.
- Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrugresistant, extensively drug-resistant and pandrugresistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. ClinMicrobiol Infect. 2012;18:268–81
- Cheesbrough M. District Laboratory Practice in Tropical Countries. 2nd edition. Cambridge University Press, UK. 2006. 178-187
- Clinical Laboratory Standard Institute, CLSI. Performance standards for antimicrobial disksusceptibility test. Fifteenth informational supplement, CLSI document M100-S15. 2011;CLSI, Wayne, PA. USA
- Ejikeugwu C, Iroha I, Orji J, et al. Antibiogram of Esbl- Producing Pseudomonas Aeruginosa Isolates Of Nosocomial Origin. Eur J Pharm Med Res. 2015; 2: 92-99
- UgwuMC, IgbokweJO, OkezieU, et al. Prevalence of ESBLs and MBLs among Escherichia coli and Klebsiellapneumoniae Isolates from a Nigerian Abattoir. J Trop Dis. 2018;6: 261.
- Rynga D, Shariff M, Deb M. Phenotypic and molecularcharacterization of clinical isolatesofAcinetobacterbaumannii isolated from Delhi, India. Ann ClinMicrobiolAntimicrob. 2015;14:40
- Ejikeugwu C, Esimone C, IrohaI, et al. Genotypic and Phenotypic Characterization of MBL Genes in Pseudomonas aeruginosa Isolates from the Non-hospital Environment. J Pure Appl Microbiol. 2018; 12: 1877-85.
- Alaka OO, Orimolade EA, Ojo OO, et al. The Phenotypic Detection of Carbapenem Resistant Organisms inOrthopaedic Wound nfections in Ile-Ife, Nigeria. Acta Scientific Microbiol. 2019; 2: 35-42.
- Paterson DL, Bonomo RA. Extended-spectrum β- lactamases: a clinical update. ClinMicrob Rev. 2005;18:657-86
- Gefen Halevi S. Isolation of genetically unrelated bla (NDM-1)-positive Providencia rettgeri strains in Israel. J ClinMicrobiol. 2013; 51:1642–43.
- Jena J, Sahoo RK, Subudhi E, et al. Prevalence of ESBL, MBL and AmpC â-Lactamase producing multidrug resistance gram negative bacteriaTertiary Care Hospital. J Pure Appl Microbio. 2014;8: 4099-105.
- Stu¨renburg E, Mack D. Extended-spectrum lactamases: implications for the clinical microbiology laboratory, therapy, and infection control. J Infect. 2003; 47:273–95.
- Lincopan N, Renato L, Marco A, et al. Enterobacteria producing extended-spectrum b-lactamases andIMP-1 metallo-b-lactamases isolated from Brazilian hospitals. J Med Microbiol. 2006; 55:1611-13.
- Ibadin EE, Omoregie R, Igbarumah IO, et al. Prevalence of extended spectrum b-Lactamase, AmpC b-Lactamase and Metallo-b-Lactamase among gram negative bacilli recovered from clinical specimens in Benin city, Nigeria. Int J Enteric Pathog. 2017;5: 85-91
- Dropa M, Balsalobre LC, Lincopan N, et al. Extendedspectrum beta-lactamases among Enterobacteriaceae isolated in a public hospital in Brazil. Rev Inst Med Trop S Paulo. 2009; 51: 203-09.