Research Article - Asian Journal of Biomedical and Pharmaceutical Sciences (2019) Volume 9, Issue 67
Docking study and result conclusion of heterocyclic derivatives having urea and acyl moiety.
Rakhi Mishra1*, Avijit Mazumder1, Rupa Mazumder1, Prem Shankar Mishra2 and Preeti Chaudhary3
1Department of Pharmaceutical Chemistry, Noida Institute of Engineering & Technology (Pharmacy Institute), Greater Noida, India
2Department of Pharmacy, Galgotias University, Greater Noida, India
3Department of Pharmacy, Meerut Institute of Engineering and Technology, Meerut, Uttar Pradesh, India
- *Corresponding Author:
- Rakhi Mishra
Department of Pharmaceutical Chemistry
Noida Institute of Engineering & Technology (Pharmacy Institute)
Greater Noida, India
Accepted date: May 02, 2019
Citation: Rakhi Mishra et.al. Docking study and result conclusion of heterocyclic derivatives having urea and acyl moiety. Asian J Biomed Pharmaceut Sci. 2019;9(67):13-17.
DOI: 10.35841/2249-622X.67.19-082
Visit for more related articles at Asian Journal of Biomedical and Pharmaceutical SciencesAbstract
In the field of molecular modeling, Docking is a method which predicts the preferred orientation of one molecule to a second when bound to each other to form a stable complex. Knowledge of the preferred orientation in turn may be used to predict the strength of association or binding affinity between two molecules. In this study a series of synthesized compounds were evaluated for focusing on binding modes, potential interactions and specific binding sites. Chemically compounds bear both the moiety acyl as well as urea and their interaction by using in silico study was investigated with beta tubulin protein that interferes with the tubulin-microtubule equilibrium, crucial for cellular mitosis. In silico studies revealed that synthesized and tested compounds show 1-7 no. of interactions with amino acids of tubulin protein. Docking study was done by using Autodock, it was found that among different synthesized compound some shows the highest and best scoring pose (lowest energy) which was -2.94 Kcal/mole between N (11) and Leucine (113) and -3.09 between N (11) and Alanine (149). Compounds with amino, hydroxy, methyl, methoxy, trimethoxy and dimethoxy substitution in derivatives of acyl urea gave an idea that urea and acyl when used in combination in different synthesized compounds show good antitumor activity.
Keywords
Docking, Urea, Acyl, Tubulin, Autodock
Introduction
Computer-aided docking is an important tool for gaining understanding of the binding interactions between a ligand and its target receptor (mainly, a protein). Multistep docking strategies have also been employed for flexible ligand docking [1-5]. Some common docking programs that utilize such fragment-based approaches include Molegro, Auto Dock, LUDI, FlexX, Growmol, Hook, Q-fit and Surflex [6-8] (Figure 1).
Figure 1: Docking of ligand and recptor [9].
It was therefore considered of interest to synthesize a variety of cinnamic acid derivatives wherein substituted cinnamoyl chlorides were reacted with urea to obtain acyl urea’s. These new compounds possess features similar to above mentioned series of compounds and are expected to exhibit similar pattern of pharmacological activities which in this case are anti-tumor activity sedative- hypnotic, smooth muscle relaxant activities.
Methodology
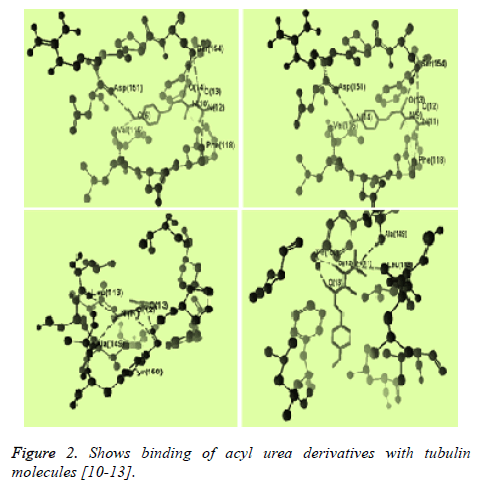
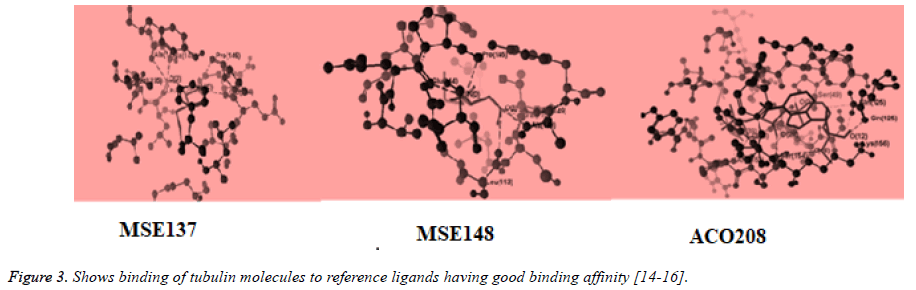
The structure of tubulin was obtained from the Protein Data Bank (www.pdb.org). Compound structures were built with ChemBio Draw Ultra 11.0 and their geometry optimization was performed using the MMFF94 method in the program and saved as MRV format [9,10]. The test compound were placed into binding site of tubulin molecule by import method to the reference ligand ACO201(A), MES(137), MES(148) which are selective inhibitor against tubulin The binding affinity between ligand (synthesized compounds) and enzyme (docking score) was predicted using MolDock Score, plant score using algorithm of iterated complex and mole dock optimizer [11-15] (Figures 2 and 3).
STEP 1 - Auto Dock / Molegro software was opened
Clicked on file
• Molecule of protein taken from protein database was imported
• Cofactor water, ligands were removed
• Protein molecule was imported
Clicked on preparation
• Prepared molecule of protein
• Detected cavities than click on OK
Again clicked on file
• Followed same step as A but only import ligand now
Again repeated step B of preparation
Clicked on file, saved workspace
Clicked on docking
Docking wizard was opened
Ligands were chosen
Plant score, mole dock score was used using mole dock complex algorithm
Results were obtained and saved
Result and Discussion
Newly synthesized acyl urea derivatives were evaluated for their antitumor activity by using Autodock software program. By using Autodock software values of different parameters like no of interactions, bond length and bond energy were obtained. All the measured values are compared with three reference compounds.
In the result it was found that some acyl urea derivatives are having bond energy and bond length which was of significance when compared with reference compounds used such as ACO201(A), MES(137) and MES(148). It was found that compound 1a (acyl urea derivative) have binding energy of -0.27589 Kcal/mole which was similar and comparable to reference compounds binding energy with tubulin protein. It was also observed in the result that in all acyl urea derivatives, nitrogen atom of urea have the best binding energy score when checked for binding with tubulin protein. All the compounds are having 4-6 no of interactions which reflects that the binding poses of the docked acyl urea compound ensures favourable and valid potential binding modes with tubulin protein
| Docking of protein (Tubulin) with ligands | ||||||
|---|---|---|---|---|---|---|
| S. No | Compound Name | No of Interactions | Protein residue name | Compound residue name | Bond Length | Bond Energy |
| (A°) | (Kcal/mole) | |||||
| 1 | 1a | 6 | Phe(118) | N(12) | 2.86065 | -2.5 |
| Ser(154) | O(14) | 2.78873 | -2.5 | |||
| Phe(118) | N(10) | 3.25137 | -0.2759 | |||
| Ser(154) | O(13) | 3.09957 | -2.5 | |||
| Asp(151) | O(6) | 3.10001 | -2.5 | |||
| Val(115) | O(6) | 3.10001 | -2.4999 | |||
| 2 | 2a | 6 | Phe(118) | N(11) | 2.88065 | -2.5 |
| Ser(154) | O(12) | 3.09991 | -2.5 | |||
| Phe(118) | N(9) | 3.25537 | 0.26664 | |||
| Ser(154) | O(13) | 2.79341 | -2.5 | |||
| Asp(151) | N(14) | 3.10011 | -2.4995 | |||
| Val(115) | N(14) | 3.10008 | -2.4996 | |||
| 3 | 3a | 4 | Tyr(150) | O(13) | 2.9897 | -7.5 |
| Tyr(150) | O(12) | 3.13828 | -7.4008 | |||
| Leu(113) | N(11) | 2.93669 | -7.1 | |||
| Ala(149) | N(11) | 3.09969 | -7.2 | |||
| 4 | 4a | 4 | Tyr(150) | O(13) | 2.9709 | -2.5 |
| Tyr(150) | O(12) | 3.12009 | -7.3995 | |||
| Leu(113) | N(11) | 2.94296 | -2.943 | |||
| Ala(149) | N(11) | 3.09964 | -3.0996 | |||
| a) | ACO201(A) | 8 | Arg(152) | N(39) | 3.34554 | -0.6456 |
| Arg(152) | O(36) | 2.84055 | -2.5 | |||
| Gln(53) | N(39) | 3.57519 | 0.37031 | |||
| Gln(53) | O(36) | 3.0643 | -2.5 | |||
| Ser(49) | O(24) | 2.40575 | -0.8813 | |||
| Ser(154) | O(16) | 3.22297 | -1.8852 | |||
| Lys(156) | O(12) | 1.96771 | -2.8895 | |||
| Gln(125) | O(12) | 2.96183 | -2.3273 | |||
| b) | MES(137) | 5 | Glu(144) | N(0) | 3.07233 | -2.5 |
| Pro(145) | N(0) | 3.10014 | -2.4993 | |||
| Val(115) | O(3) | 3.26753 | -0.7544 | |||
| Leu(113) | O(3) | 2.83785 | -2.5 | |||
| Ala(149) | O(3) | 3.10025 | -2.4988 | |||
| Ala(149) | O(3) | 2.62258 | -2.5 | |||
| c) | MES(148) | 6 | Glu(144) | N(0) | 3.09915 | -2.5 |
| Pro(145) | N(0) | 3.08851 | -2.5 | |||
| Val(115) | O(3) | 2.87091 | -2.5 | |||
| Leu(113) | O(3) | 3.25601 | -0.7973 | |||
| Ala(149) | O(3) | 3.10042 | -2.4979 | |||
| Ala(149) | O(3) | 2.59975 | -2.4979 | |||
Table 1: Binding affinity between ligand and tubulin protein [17].
Conclusion
The molecular docking study of acyl urea derivatives with tubulin protein revealed that acyl urea’sx are having good interaction in favourable pose with tubulin which was explained by lowest binding energy, strong bond length and 4-6 no of interactions with active site of tubulin molecule Thus it can be concluded that some acyl urea derivatives could be used as a template for the future development through modification or derivatization to design more potent therapeutic agents. Compounds synthesized if properly changed into therapeutic agent can be used for antitumor action.
Acknowledgement
We are really thankful to the management of Noida Institute of Engineering and Technology, Greater Noida for providing facilities of research work and granting us a chance to present our work in front of you.
References
- Meng EC, Shoichet BK, Kuntz ID. Automated docking with grid-based energy evaluation. J Comput Chem 1992; 13:505–524.
- Jorgensen WL. The many roles of computation in drug discovery. Science. 2004; 303: 1813–1818.
- Kitchen DB, Decornez H, Furr JR, et al. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov. 2004; 3: 935–949.
- McConkey BJ, Sobolev V, Edelman M. The performance of current methods in ligand-protein docking. Current Science. 2002; 83: 845–855.
- Koshland DE Jr. Correlation of structure and function in enzyme action. Science. 1963; 142: 1533–1541.
- Leach AR, Kuntz ID. Conformational analysis of flexible ligands in macromolecular receptor sites. J Comput Chem 1992; 13: 730–748.
- Head RD, Smythe ML, Oprea TI, et al. Validate: A new method for the receptor-based prediction of binding affinities of novel ligands. J Am Chem Soc 1996; 118: 3959–3969.
- Meng EC, Shoichet BK, Kuntz ID. Automated docking with grid-based energy evaluation. J Comput Chem 1992; 13: 505–524.
- Sanphanya K, Wattanapitayakul SK, Prangsaengtong O. Synthesis and evaluation of 1-(substituted)-3-prop-2-ynylureas as antiangiogenic agents: Bioorg Med Chem Lett 2012; 22: 3001-3005.
- Glaser F, Morris RJ, Najmanovich RJ. A method for localizing ligand binding pockets in protein structures. Proteins. 2006; 62: 479–488.
- Diller DJ, Merz KM Jr. High throughput docking for library design and library prioritization. Proteins. 2001; 43: 113–124.
- Lorber DM, Shoichet BK. Flexible ligand docking using conformational ensembles. Protein Sci. 1998; 7: 938–950.
- Feher M, Deretey E, Roy S. BHB: A simple knowledge-based scoring function to improve the efficiency of database screening. J Chem Inf Comput Sci. 2003; 43: 1316–1327.
- Eldridge MD, Murray CW, Auton TR. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput Aided Mol Des. 1997; 11: 425–445.
- Muegge I, Martin YC. A general and fast scoring function for protein-ligand interactions: A simplified potential approach. J Med Chem. 1999; 42: 791–804.
- Charifson PS, Corkery JJ, Murcko MA, et al. Consensus scoring: A method for obtaining improved hit rates from docking databases of three-dimensional structures into proteins. J Med Chem. 1999; 42: 5100–5109.
- Gschwend DA, Kuntz ID. Orientational sampling and rigid-body minimization in molecular docking revisited: On-the-fly optimization and degeneracy removal. J Comput Aided Mol Des. 1996; 10: 123–132.