Research Article - Virology Research Journal (2017) Virology Research Journal (Special Issue 1-2017)
Molecular characterization of a Begomovirus and Betasatellite infecting wild sunflower (Helianthus spp.) in India
Susheel Kumar, Meraj Jaidi, Raj SK*Plant Molecular Virology Laboratory, Council of Scientific and Industrial Research-National Botanical Research Institute, Lucknow, UP, India
- Corresponding Author:
- Raj SK
Plant Molecular Virology Laboratory
Council of Scientific and Industrial Research
National Botanical Research Institute
Lucknow 226001, UP
India
Tel: 091-522-2297950
Fax: 091-522-2205836
E-mail: skraj2@rediffmail.com
Accepted Date: December 29, 2016
Citation: Kumar S, Jaidi M, Raj SK. Molecular characterization of a Begomovirus and Betasatellite infecting wild sunflower (Helianthus spp.) in India. Virol Res J. 2016;1(1):1-9.
Copyright: © 2016 Mbamalu ON, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Wild sunflower (Helianthus spp.) plants exhibiting Begomovirus disease like symptoms: yellow vein net and leaf curl were observed growing in road side in Jaipur (Rajasthan), India. The causal virus was successfully transmitted on healthy seedlings of Helianthus spp. by whitefly (Bemisia tabacci) which induced similar symptoms to those of naturally infected plants that indicated presence of Begomovirus. The association of Begomovirus and betasatellite with these symptoms of Helianthus spp. was investigated by sequence analyses of viral DNA genome and satellite DNAs amplified by rolling circle amplification (RCA) and polymerase chain reaction (PCR), respectively. The highest sequence identities and closest phylogenetic relationships for the Begomovirus and betasatellite detected in Helianthus spp. were to Ageratum enation virus (AEV) and Ageratum leaf curl betasatellite (ALCB), respectively. These findings identified the virus and satellite DNAs infecting Helianthus spp. as AEV and ALCB, respectively. Helianthus spp. and Tobacco (Nicotiana glutinosa) seedlings infected with cloned AEV and ALCB by Agroinoculation developed yellow vein net and leaf curl symptoms, whereas plants infected with AEV alone or ALCB did not develop any symptom. The results show that this complex infects and causes disease in Helianthus spp. and Tobacco. Helianthus spp. is an invasive weed commonly found around agricultural fields and along roadsides in Rajasthan, India. These results also indicate that Helianthus spp. plants infected with AEV and associated betasatellite DNA may act as an alternate host (reservoir) for other economically important plants.
Keywords
Ageratum enation virus, Ageratum leaf curl betasatellite, Agroinfectivity assay, Wild sunflower, Yellow vein net and leaf curl disease.
Introduction
Wild sunflower (Helianthus spp.) belongs to family Asteraceae which grows as weed in roadsides, fields, along desert washes and in grassy areas. This weed is drought-resistant and tolerant to various growth conditions. It causes the greatest harm in summer grain cereals because of the showy sunflower sprouts simultaneously with summer wheat, overtaking the latter in growth, drying and exhausting soil, which results in significant reduction in crop yield; mass weediness results in complete destruction of crops (http://www.agroatlas.ru/en/content/ weeds/Helianthus_lenticularis/). It is also reported to be used for broadening the genetic base of cultivated sunflower in India and to introgress the desirable traits from wild sunflowers to cultivated sunflower [1].
Literature survey revealed that none of the plant virus has been reported on wild sunflower (Helianthus spp.) worldwide except an unpublished record of sequence data of Tobacco curly shoot virus (Accession HQ407395) and Ageratum yellow leaf curl betasatellite (Accession HQ407397) molecules infecting wild sunflower (Helianthus spp.) in India are available in GenBank database. Tomato leaf curl Karnataka virus and Potato apical leaf curl betasatellite associated with leaf curl disease of sunflower (Helianthus annuus) has been reported from Karnataka state of India [2]. Partial sequence data on Ageratum enation virus (AEV) (Verbesina encelioides yellow vein Lakshmangarh virus) isolate (JN998449) and Ageratum leaf curl betasatellite (Verbesina yellow vein betasatellite) isolate (JQ693145) infecting wild sunflower (Verbesina encelioides) are also available from India in GenBank database. However, we report the natural occurrence of Ageratum enation virus (AEV) and Ageratum leaf curl betasatellite (ALCuB) infecting wild sunflower (Helianthus spp.) plants.
Viruses in the family Geminiviridae have circular single-stranded (ss) genomes encapsidated in twinned icosahedral (geminate) virions. Gemini viruses transmitted by the whitefly Bemisia tabaci (Gennadius) are in the genus Begomovirus and infect a wide range of crops including tomato [3,4]. Begomoviruses have either a monopartite or bipartite genome organization [5]. Monopartite Begomoviruses have genomes consisting of a single component that encode six genes: two in the virion sense, V1 (capsid protein) and V2 (pre-capsid protein); and four in the complementary sense, C1 (replication-associated protein), C2 (transcription activator protein), C3 (replication enhancer protein) and C4 (C4 protein). Monopartite Begomoviruses may associate ~1.4 kb (half the genome size of the helper Begomovirus) ssDNA molecules known as betasatellites and alphasatellites [6-8]. Betasatellites are symptom-modulating and are widespread across the Old World [9]. The βC1 gene encoded by the betasatellites has important roles in symptom induction, in suppression of transcriptional and post transcriptional gene silencing, and can affect jasmonic acid responsive genes [10].
Alphasatellites are self-replicating satellite-like molecules that are dependent on a helper virus for movement in and between plants. Alphasatellites occur with monopartite Begomoviruses and betasatellites in the Old World [4,11] and with bipartite Begomoviruses (but no betasatellites) in the New World [12]. The precise function of alphasatellites is unclear [10]. However, Nawaz-Ul-Rehman showed that alphasatellites may be involved in overcoming RNA-mediated host defense and attenuating symptoms of the helper virus [13]. The work of Nawaz-Ul- Rehman also showed that some alphasatellites suppress PTGS (host defense) [13].
In this communication we report the molecular characterization of a Begomovirus (AEV) together with Ageratum leaf curl betasatellite (ALCuB) infecting wild sunflower (Helianthus spp.) plants and causing yellow vein net and leaf curl symptoms. We further confirmed that the cloned DNAs of AEV and ALCuB cause yellow vein net and leaf curl disease in Helianthus spp.
Materials and Methods
Sample collection and whitefly transmission
Leaf samples were collected from five diseased plants showing yellow vein net and leaf curl symptoms and a healthy plant of wild sunflower (Helianthus spp.) growing near road side in Jaipur (Rajasthan), India during December 2015. The whitefly transmission test was attempted using B biotype whiteflies (Bemisia tabaci). The adult B. tabaci, starved for 2 h, were allowed to feed on infected Helianthus spp. for a 24-h acquisition period, then transferred onto healthy Helianthus spp. seedlings (Ten B. tabaci per seedling) and allowed a 24-h inoculation access period. The inoculated plats were maintained in insect proof glass house for about 35 days post inoculation for symptom development if any.
DNA isolation, nucleic acid spot hybridization (NASH) assays and PCR
The total DNA was isolated from 100 mg leaf tissue of five naturally infected as well as a healthy and whitefly transmitted Helianthus spp. plants using a DNeasy Plant Mini Kit (Qiagen GmbH, Hilden, Germany). For detection of Begomovirus infection, nucleic acid spot hybridization assays (NASH) was done using a radiolabelled probe generated from a Begomovirus clone [14].
Further, the PCR was performed with Begomovirus specific degenerate primers, PALIv1978 and PARIc496 following the described conditions [15]. All these samples were also checked for the presence of a Begomovirus DNA B component with the primers BC1F and BC1R [16] and betasatellite DNA with primers Beta 01 and Beta 02 [7] following the recommended conditions. All PCR products were analyzed by electrophoresis in 1.0% agarose gels with the 1 kb DNA ladder (Thermo Fisher Scientific Inc., Pittsburgh, USA) as size markers.
Full length viral genome amplification, cloning and sequencing of amplicons
The full-length Begomovirus genome was amplified from the DNA isolated from fifteen symptomatic samples by the rolling circle amplification (RCA) method using an Illustra TempliPhi Amplification kit (GE Healthcare, Buckinghamshire, UK). The resulting products were digested with BamHI, XbaI, DraI and XhoI and analyzed by electrophoresis in 1.5% agarose gels to identify enzymes to clone full length genomic DNA. Putative full-length (~2.7 kb) amplicons from three randomly selected samples were cloned into the pCAMBIA1300 vector digested with the appropriate enzyme. The cloned Begomovirus genome (~2.7 kb) and ~1.3 kb DNA fragments obtained by PCR using and betasatellite-specific primers were sequenced and deposited in GenBank database under accession numbers KY089033 and KY089034, respectively.
Sequence analyses
The sequences were compared with those publicly available in the NCBI sequence database by BLASTn analysis for sequence identities. The open reading frames (ORFs) located in the genome and their predicted amino acid sequences were analyzed with the ORF finder (www.ncbi.nlm.nih.gov/ projects/gorf/) and ExPasy (http://www.expasy.org/resources/ search/keywords:translation) tools, respectively. The species demarcation tool (SDT, Muhire et al. 2014) was used with default parameters to obtain the specific nucleotide sequence identity of Begomovirus, alphasatellite and betasatellites identified here with respective sequences of DNA molecules. The amino acid identity of protein coding ORFs of these molecules were obtained by the DiAlign tool (http://www.genomatix.de/cgi-bin/ dialign/ dialign.pl). The evolutionary history was inferred using the Maximum Parsimony method and the tree was obtained using the Close-Neighbor-Interchange algorithm in MEGA v6.1 program [17].
Construct preparation and Agro-infectivity assays
Constructs for the infectivity of the Begomovirus were produced by digesting the clone with BamHI and HindIII and ligating the resulting 2,607 bp fragment into the pCAMBIA1300 vector. Then, the full-length genomic 2746 bp BamHI fragment was ligated at the BamHI site in the sense orientation. For preparation of infectious clone of the betasatellite, the RCA amplified DNAs were partially digested with SmaI and EcoRI, respectively which produced ~1.4 kb monomer and ~2.8 kb dimer. The dimer of ~2.8 kb recovered from both the digestions were eluted independently and ligated at respective cloning site of pCAMBIA1300 vector. The constructs for the infectivity of the Begomovirus and betasatellite will henceforth be referred to as SKJA-Agro and SKJB-Agro respectively. pCAMBIA1300 constructs were mobilized into Agrobacterium tumefaciens (GV3101) and Agroinoculated by the syringe infiltration method (in combination or separately) into leaf lamina of Helianthus spp. and Nicotiana glutinosa seedlings at the 4 to 6 leaf stage. The infiltrated plants were kept in glasshouse conditions (natural illumination with 25°C ± 2°C temperature) and examined regularly for appearance of symptoms. The experiments were repeated three times with ten plants of each species for each inoculum.
Results
Disease symptoms and virus transmission
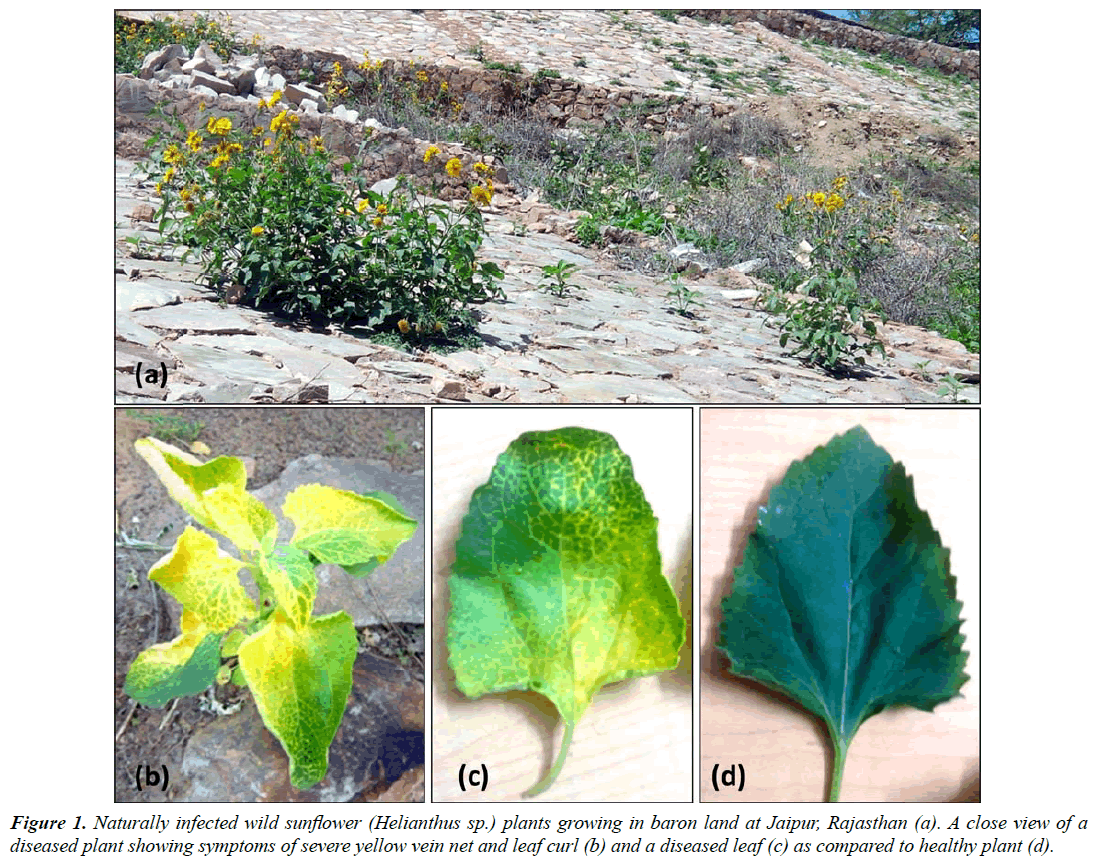
The golden yellow mosaic disease was noticed on a number of wild sunflower (Helianthus sp.) plants growing in road side in Jaipur (Rajasthan), India during December 2015 with the disease incidence about 60% (Figure1a). The infected plants exhibited golden yellows accompanied with yellow vein net on leaf and leaf curl symptoms (Figure 1b and 1c) as compared to healthy (Figure 1d) ones. The disease symptoms in these plants were similar to those in other plants induced by Begomoviruses.
About 4-6 whiteflies (Bemisia tabaci) per plant were also observed on Helianthus spp. plants (with and without symptoms). Based on presence of a whitefly population and typical Begomovirus-like (yellow vein net and leaf curl) symptoms, the causal agent was suspected to be a Begomovirus. Therefore, a whitefly transmission test was attempted onto healthy Helianthus spp. seedlings using B. tabaci. As a result, the causal pathogen was successfully transmitted from naturally infected to healthy Helianthus spp. seedlings which developed similar yellow vein net and leaf curl symptoms (5/5 plants) at 25-30 days post inoculation (dpi), suggesting Koch’s postulates.
Begomovirus detection by NASH assays and PCR
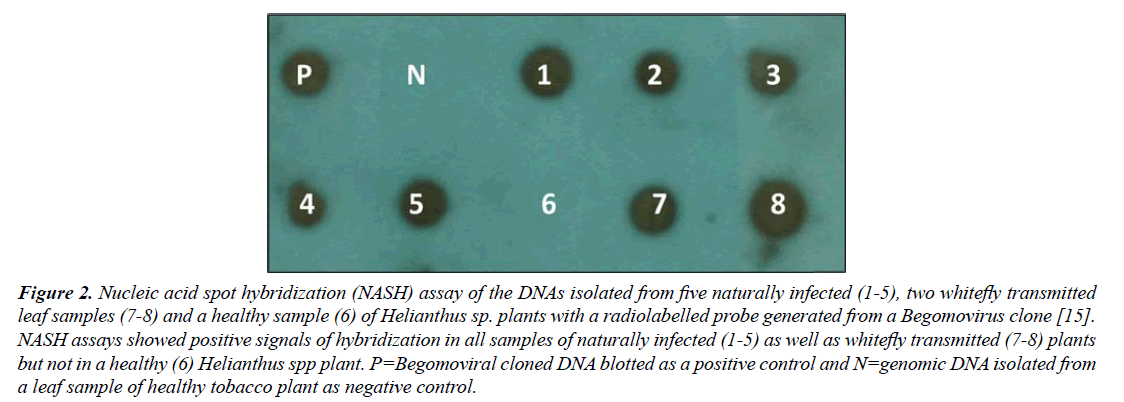
NASH resulted in positive signals of hybridization with a Begomovirus probe in all 5/5 samples of naturally infected as well as in all 2/2 whitefly-transmitted Helianthus spp. plants, similar to a positive control, while no signals of hybridization was obtained in a healthy plant (Figure 2). These results indicated the presence of Begomovirus.
Figure 2: Nucleic acid spot hybridization (NASH) assay of the DNAs isolated from five naturally infected (1-5), two whitefly transmitted leaf samples (7-8) and a healthy sample (6) of Helianthus sp. plants with a radiolabelled probe generated from a Begomovirus clone [15]. NASH assays showed positive signals of hybridization in all samples of naturally infected (1-5) as well as whitefly transmitted (7-8) plants but not in a healthy (6) Helianthus spp plant. P=Begomoviral cloned DNA blotted as a positive control and N=genomic DNA isolated from a leaf sample of healthy tobacco plant as negative control.
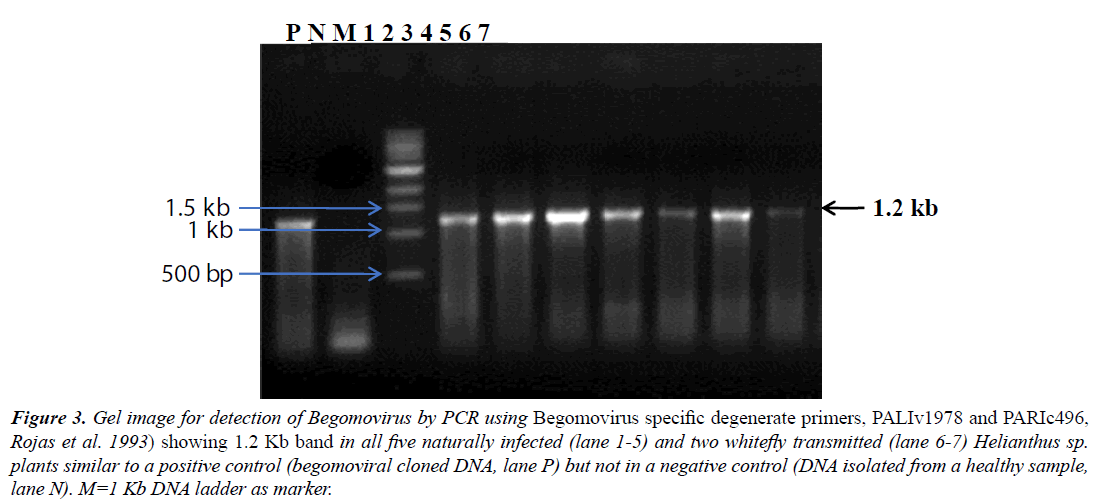
The Begomovirus infection was further confirmed by PCR performed using Begomovirus specific degenerate primers (PALIv1978 and PARIc496, [15], which resulted in amplification of ~1.2 kb DNA fragments from all 5/5 samples of naturally infected and 2/2 whitefly transmitted plants of Helianthus sp. suggesting Begomovirus infection, whereas no fragment was amplified from a non-symptomatic plant (Figure 3). All samples were also assessed for the presence of a DNA-B component by PCR with specific primers. Because no DNA fragments were amplified from any of seven samples, this suggested the Begomovirus was monopartite.
Figure 3: Gel image for detection of Begomovirus by PCR using Begomovirus specific degenerate primers, PALIv1978 and PARIc496, Rojas et al. 1993) showing 1.2 Kb band in all five naturally infected (lane 1-5) and two whitefly transmitted (lane 6-7) Helianthus sp. plants similar to a positive control (begomoviral cloned DNA, lane P) but not in a negative control (DNA isolated from a healthy sample, lane N). M=1 Kb DNA ladder as marker.
Characterization of the Begomovirus associated with Helianthus spp.
RCA products obtained from five naturally infected symptomatic leaf samples were digested with BamHI, XbaI, DraI and XhoI. The expected size ~2.7 kb DNA fragment was observed in all five samples following digestion with BamHI only. The ~2.7 kb fragment from three positive samples were cloned and sequenced. The sequences of three isolates were identical to each other; therefore, the 2746 nucleotide sequence of one isolate (from Helianthus spp.) was deposited in the GenBank database under accession number (KY089033). Analysis of the sequence revealed that the Begomovirus characterized here contains six open reading frames: V1 and V2 in the virion sense and C1, C2, C3 and C4 in the complementary sense and an intergenic region. The Begomovirus isolated from Helianthus spp was designated as SKJA isolate.
During BLASTn analysis, the virus sequence (KY089033) isolated from Helianthus spp. (SKJA isolate) shared 95% to 98% with complete genome of Ageratum enation virus (AEV) isolates: AM698011 (Ageratum conyzoides-Pakistan); KC795968 (Solanum lycopersicum-India); AM701770 (Brassica rapa-Pakistan); AJ437618 (Ageratum conyzoides- Nepal); GQ268327 (Trichosanthes dioica-India); FJ177031 (Cleome gynandra-India); EU867513 (Amaranthus cruentus- India). It also shared 92% to 93% identities with AEV isolates: KP195264 (Solanum lycopersicum-India); KJ488991 (Ageratum houstonianum-India); FN794201 (Crassocephalum crepidioides-India) [18]; KM262822 (Calendula officinalis- India); KJ488990 (Ageratum houstonianum-India); JQ911767 (Ageratum sp. (ornamental)-India); JX436472 (Solanum lycopersicum-India). While identities were scored as 88% to 89% with Tobacco curly shoot virus isolates: HQ407395 (Helianthus spp.- India); KM383757 (Solanum lycopersicum- Bangladesh); GU001879 and AJ420318 (pepper-China) [19] and 87% with Tomato leaf curl Bangladesh virus isolates: KM383762, KM383765 and KM383759 (Solanum lycopersicum-Bangladesh).
Pairwise sequence comparison of nucleotide sequence of the DNA genome and amino acid sequences of various ORFs of the SKJA isolate with respective sequences of various isolates of AEV (appeared in BLASTn) was also performed (Table 1). The SKJA isolate shared the highest nucleotide sequence identity (98.3%) with three isolates of AEV (KM262822, GQ268327 and JF682242) reported from India and between 95.7% and 97.5% nucleotide sequence identity with other AEV isolates reported from India and Pakistan (Table 1). The coat protein (V1) gene of the virus isolate also showed 98.4% to 100% amino acid sequence identities with respective sequences of various isolates of AEV considered for study. The other genes: V1, C1, C2, C3 and C4 of the virus isolate under study shared 46.8% to 88.6%, 75.3% to 93.1%, 88.1% to 92.5%, 88.1% to 100% and 52.6% to 100%, respectively (Table 1).
| Accessions | Virus | Host | Location | DNA genome | V2 | V1 | C3 | C2 | C1 | C4 |
|---|---|---|---|---|---|---|---|---|---|---|
| KM262822 | AEV | Calendula officinalis | India | 98.3 | 46.8 | 98.8 | 89.5 | 91.0 | 92.2 | 95.3 |
| GQ268327 | AEV | Amaranthus cruentus | India | 98.3 | 87.8 | 98.8 | 88.8 | 90.3 | 91.4 | 82.4 |
| JF682242 | AEV | Solanum lycopersicum | India | 98.3 | 85.3 | 97.3 | 88.1 | 89.6 | 92.2 | 82.1 |
| KP195264 | AEV | Solanum lycopersicum | India | 97.5 | 85.1 | 99.2 | 88.1 | 88.1 | 92.2 | 84.7 |
| KJ488991 | AEV | Brassica rapa | India | 97.5 | 85.8 | 98.8 | 100.0 | 92.5 | 92.2 | 92.9 |
| KC795968 | AEV | Solanum lycopersicum | India | 97.5 | 86.4 | 100.0 | 90.3 | 92.5 | 75.3 | 52.6 |
| JX436472 | AEV | Ageratum sp. | India | 97.5 | 85.7 | 99.2 | 100.0 | 92.5 | 92.8 | 83.5 |
| JQ911767 | AEV | Ageratum houstonianum | India | 97.5 | 86.0 | 99.2 | 90.3 | 92.5 | 92.2 | 96.5 |
| KJ488990 | AEV | Ageratum houstonianum | India | 97.5 | 86.1 | 99.2 | 89.6 | 92.5 | 92.2 | 96.5 |
| KJ488991 | AEV | Brassica rapa | India | 97.5 | 85.8 | 98.8 | 100.0 | 92.5 | 92.2 | 92.9 |
| FJ177031 | AEV | Cleome gynandra | India | 97.4 | 88.1 | 98.8 | 92.5 | 89.6 | 92.5 | 81.2 |
| AM698011 | AEV | Ageratum conyzoides | Pakistan | 97.4 | 86.4 | 98.4 | 88.1 | 88.8 | 90.6 | 82.4 |
| JQ911765 | AEV | Papaver somniferum | India | 97.4 | 85.1 | 98.8 | 88.6 | 89.6 | 92.0 | 81.2 |
| FN794201 | AEV | Crassocephalum crepidioides | India | 96.7 | 85.5 | 99.2 | 90.3 | 91.1 | 93.1 | 94.1 |
| AJ437618 | AEV | Ageratum conyzoides | India | 96.6 | 87.0 | 99.2 | 100.0 | 88.1 | 92.2 | 80.0 |
| EU867513 | AEV | Amaranthus hypochondriacus | India | 96.6 | 88.6 | 98.4 | 88.1 | 89.6 | 91.4 | 81.2 |
| AM701770 | AEV | Ageratum conyzoides | India | 95.7 | 87.2 | 98.4 | 100.0 | 90.3 | 92.0 | 100.0 |
Table 1: Sequence identities of Ageratum enation virus of Helianthus sp. with other selected Begomovirus genomes at nucleotide (nt) and their ORFs at amino acid (aa) levels.
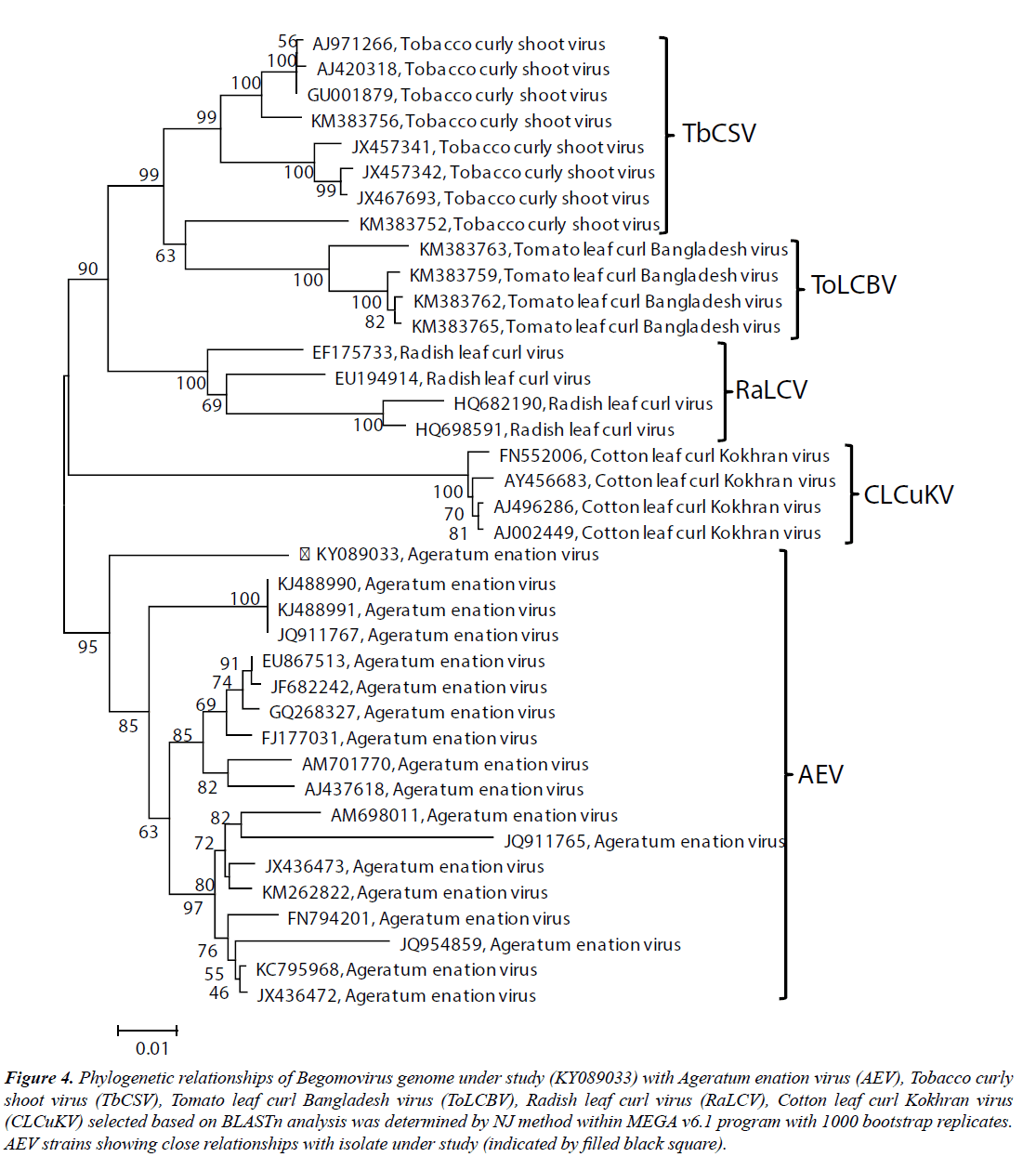
To determine the phylogenetic relationship of the SKJA isolate with other close Begomoviruses, phylogeny was done using the MEGA v6.1 program. The SKJA isolate showed closest relationship with isolates of Ageratum enation virus (AEV): KJ488990, KJ488991 (Ageratum houstonianum) and JQ911767 (Ageratum sp., ornamental) reported from India and close relationships with other AEV isolates considered for this study (Figure 4). Based on high sequence identity and close phylogenetic relationships, and Begomovirus species demarcation criteria of >91% identity recommended by the ICTV Geminiviridae study group (Brown et al. 2015), the Begomovirus isolate SKJA was identified as an isolate of AEV for which the isolate descriptor AEV-[In:Lko:Hel:16] is proposed.
Figure 4: Phylogenetic relationships of Begomovirus genome under study (KY089033) with Ageratum enation virus (AEV), Tobacco curly shoot virus (TbCSV), Tomato leaf curl Bangladesh virus (ToLCBV), Radish leaf curl virus (RaLCV), Cotton leaf curl Kokhran virus (CLCuKV) selected based on BLASTn analysis was determined by NJ method within MEGA v6.1 program with 1000 bootstrap replicates. AEV strains showing close relationships with isolate under study (indicated by filled black square).
Characterization of satellite molecules associated with Helianthus sp.
Because monopartite Begomoviruses are reported to be associated with satellites, PCR analyses with betasatellite-specific primers were performed with three DNA samples of infected Helianthus spp. (from which the Begomovirus genome was amplified). The primers directed the amplification of ~1.4 kb DNA fragments from all three infected samples which indicated the association of a betasatellite with AEV in Helianthus sp. The ~1.4 kb betasatellite components obtained from SKJ1 sample were sequenced and deposited in GenBank under accession numbers: KY089034 (SKJB-betasatellite). The betasatellite associated with Begomovirus isolated from Helianthus spp was designated as SKJB-betasatellite isolate. Sequence analysis of the SKJBisolate revealed 1366 nucleotides that encode for a conserved βC1 ORF in the complementary-sense orientation as well as the satellite conserved region (SCR) and an A-rich region.
BLASTn analysis showed that the isolate SKJB (KY089034) showed highest 97% sequence identity with Ageratum leaf curl betasatellite (ALCB) isolates: JQ408218 (Ageratum sp.), KR922823 (Calendula officinalis), KR922821 (C. officinalis), KR922822 (C. officinalis) reported from India. While the identities were 90% to 94% with other ALCB isolates: KX108998 (Fenugreek), AM412239 (Sonchus oleraceus), AM698010 (Ageratum conyzoides), AM701771 (Brassica rap), AM494977 (Digera arvensis), F728868 (Ageratum conyzoides), JF728869 (Daucus carota), KJ028213 (Solanum nigrum), KC305084 (wheat), KC305085 (wheat), HM143915 (Nicotiana glutinosa), KJ948106 (Papaver somniferum), JX512904 (Amaranthus hypochondriacus), JQ710745 (Amaranthus cruentus) and KU376492 (Calotropis procera) reported from India and Pakistan.
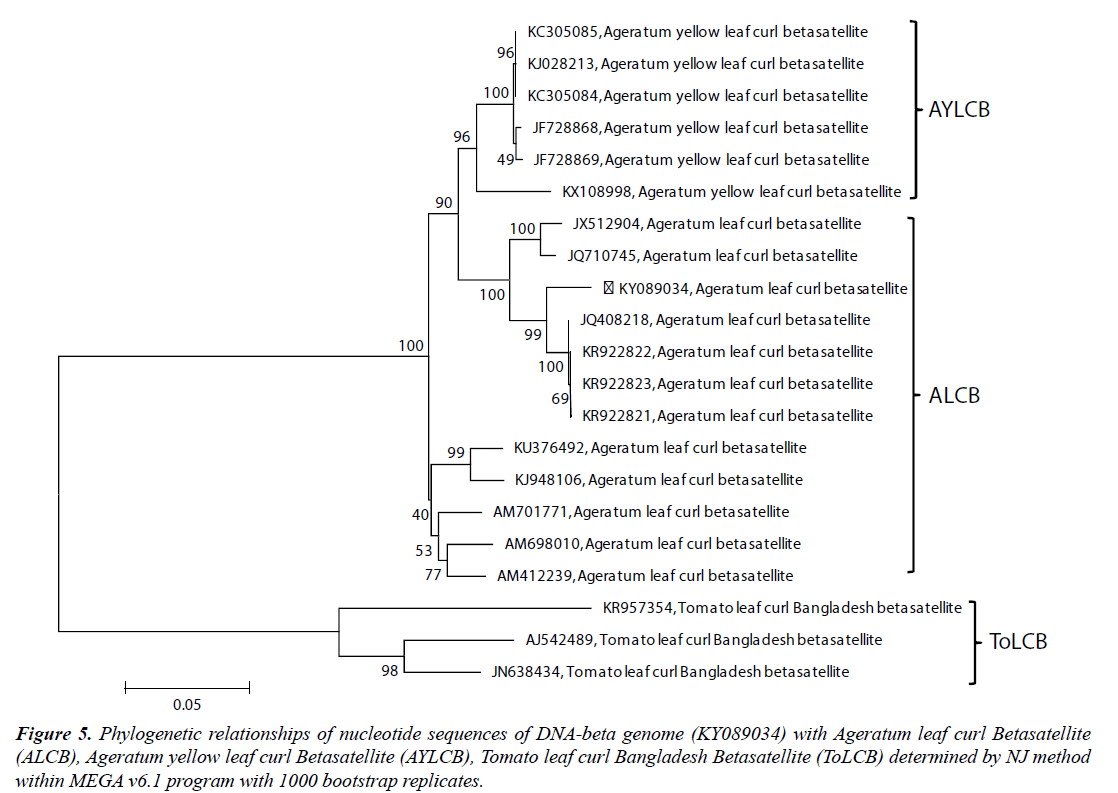
The pairwise sequence identity of the complete SKJBbetasatellite sequence (KY089034) revealed highest 97.4% nucleotide identities with Ageratum leaf curl betasatellite (ALCB) sequences (KR922821, KR922822, KR922823, JQ408218) and 91.8% to 94.3% identities with other ALCB. It also showed 91.7% to 91.8% identities with Ageratum yellow leaf curl betasatellite (AYLCB) isolates (Table 2). The βC1 ORF of the SKJB-betasatellite showed 78.90% to 79.70% amino acid sequence identities with corresponding ORF of ALCB isolates (Table 2). The phylogenetic analysis showed the close relationships of SKJB-betasatellite with isolates of ALCB and these isolates clustered together in a monophyletic group (Figure 5). Because of the high sequence identity and close relationships with ALCB, the SKJB-betasatellite was identified as isolate of ALCB for which isolate descriptor ALCB-[In:Lko:Hel:16] is proposed.
| Accessions | Betasatellite | Percent sequence identity at | |
|---|---|---|---|
| Complete nucleotide | beta C1 amino acid | ||
| KR922822 | Ageratum leaf curl beta satellite | 97.4 | 79.70 |
| KR922821 | Ageratum leaf curl beta satellite | 97.4 | 79.70 |
| KR922823 | Ageratum leaf curl beta satellite | 97.4 | 79.70 |
| JQ408218 | Ageratum leaf curl betasatellite | 97.4 | 79.70 |
| KU376492 | Ageratum leaf curl betasatellite | 94.3 | 78.90 |
| JQ710745 | Ageratum leaf curl betasatellite | 93.7 | 80.25 |
| JX512904 | Ageratum leaf curl betasatellite | 93.6 | 80.50 |
| KJ948106 | Ageratum leaf curl betasatellite | 91.9 | 78.90 |
| HM143915 | Ageratum leaf curl betasatellite | 91.8 | 78.90 |
| KC305085 | Ageratum yellow leaf curl betasatellite | 91.8 | 82.10 |
| KC305084 | Ageratum yellow leaf curl betasatellite | 91.7 | 82.10 |
| KJ028213 | Ageratum yellow leaf curl betasatellite | 91.8 | 82.10 |
Table 2: Percent sequence identities of Ageratum leaf curl betasatellite molecule of Helianthussp. with other selected betasatellite at complete nucleotide (nt) and their beta C1 ORF at amino acid (aa) levels.
Figure 5: Phylogenetic relationships of nucleotide sequences of DNA-beta genome (KY089034) with Ageratum leaf curl Betasatellite (ALCB), Ageratum yellow leaf curl Betasatellite (AYLCB), Tomato leaf curl Bangladesh Betasatellite (ToLCB) determined by NJ method within MEGA v6.1 program with 1000 bootstrap replicates.
Agro-infectivity assays
The infectivity assay was conducted with Agroinfectious clones of AEV-(In:Lko:Hel:16) and ALCB-(In:Lko:Hel:16) DNAs. Helianthus sp. and N. glutinosa plants agroinoculated with the AEV+ALCB DNAs developed prominent yellow vein and leaf curl symptoms respectively, by 35 dpi (Table 3). The symptoms that developed in experimentally infected Helianthus sp. plants were indistinguishable from the symptoms observed in Helianthus sp. plants in the field. Plants inoculated with AEV-A alone also developed mild symptoms, whereas no symptoms were observed when plants were inoculated with ALVB alone or mock (Table 3). These results satisfy Koch’s postulates for AEV with ALCB causing leaf curl disease of Helianthus sp.
| Assay plant | Agroinfectious clone/s | Symptoms (by 35 dpi) |
Symptomatic / inoculated plants | Detection of* | |
|---|---|---|---|---|---|
| DNA-A | DNA-β | ||||
| Helianthus sp. | DNA-A | Mild leaf curl | 9/10 | 9/10 | 0/10 |
| DNA-A +DNA-β | Severe leaf curl | 10/10 | 10/10 | 10/10 | |
| DNA-β | No symptoms | 0/10 | 0/10 | 0/10 | |
| N. glutinosa | DNA-A | Mild yellowing | 8/10 | 8/10 | 0/10 |
| DNA-A + DNA-β | Yellowing & leaf curl | 9/10 | 9/10 | 9/10 | |
| DNA-β | No symptoms | 0/10 | 0/10 | 0/10 | |
Table 3: Infectivity assay using seedlings ofHelianthus spp. and Nicotiana glutinosa and agroinfectious clonesofAEV-[In:Lko:Hel:16] and ALCB-[In:Lko:Hel:16] DNAs abbreviated as DNA-A and DNA-β respectively.
The presence and/or absence of AEV+ALCB DNAs in the leaves developing after inoculation of both Helianthus sp. and N. glutinosa plants were assessed by PCR with the respective specific primers. The AEV DNA was detected in newly emerged leaves of plants agroinoculated with AEV alone as well as in combinations with AEV+ALCB DNAs was also detected in Helianthus sp. and N. glutinosa plants (Table 3). These results indicated that AEV can replicate and move independently or in combinations with AEV+ALCB in inoculated plants.
Discussion
The Begomovirus species AEV was first established in 2003, when the presently applicable species demarcation criteria for the genus Begomovirus were established, based upon the sequence of the Nepal isolate (AJ437618), which is the type isolate of the species [5]. Since this time AEV has been shown to be widespread across northern India and to infect a number of distinct hosts [7]. The occurrence of AgEV has been reported on Amaranthus cruentus [14], Cleome gynandra [20], Zinnia sp. [21], Trichosanthes dioica [22], Daucus carota [23] and Solanum lycopersicum [24] from India and Brassica rapa (AM701770), Ageratum conyzoides (AM698011), Sonchus oleraceus (AM261836) from Pakistan Tahir et al. [25] are reported in literature. Tahir et al. also showed that AEV has two distinct strains, and to occur across a large geographic area [25].
It is evident from the literature that AEV has a large natural host range which includes species from the families Brassicaceae (turnip), Asteraceae (A. conyzoides, Z. elegans, C. crepidioides, Tagetes patula [17] and S. oleraceous), Amaranthaceae (A. cruentus), Cucurbitaceae (T. dioica), Apiaceae (carrot) and Cleomaceae (C. gynandra). This indicates that AEV is predominantly a weed infecting Begomovirus, although it apparently has the capacity to occasionally infect crop species and is reported to be causing losses of the minor grain crop A. cruentus [20] and carrot.
Kumar et al., Tahir et al. and Marwal et al. suggested that the common partner of AEV is the betasatellite species AYLCB [25-27]. For none of the other AEV sequences available in the databases has the associated betasatellite been identified. Of the other AYLCB isolates available in the databases, only for two have the helper Begomovirus been identified Tobacco curly shoot virus isolated from wild sunflower (Helianthus sp.) originating from India (for HQ407397) and Alternanthera yellow vein virus (AlYVV) in Sonchus arvensis originating from Pakistan (for FN432358) [26]. It is interesting to note that the AYLCB from S. arvensis was identified in co-infection with a second betasatellite, Cotton leaf curl Multan betasatellite (CLCuMB). The identification of YLCB with several distinct Begomoviruses suggests that, as has been shown for other betasatellites[28-31], this has the capacity to be trans-replicated and maintained by more than one Begomovirus [25].
Tahir et al. also suggested that the availability of infectious AEV and AYLCB clones that induce distinct symptoms in common hosts opens up the possibility of investigating the molecular basis for these differences by, for example, exchanging betasatellite components. Although initially it was isolated from weeds, they also showed that now it is crossing into cultivated species and could become a more significant problem for agriculture in the future [25].
In India, Verbesina encelioides yellow vein Lakshmangarh virus (JN998449) and Verbesina yellow vein betasatellite (JQ693145) have been reported to associated with yellow vein disease of Verbesina encelioides. However, to the best of our knowledge, the present study is the first report on the molecular characterization of AEV and Ageratum leaf curl betasatellite (ALCB) associated with yellow vein net and leaf curl disease in wild sunflower (Helianthus spp.). Since wild sunflower (Helianthus spp.) is found grows as an invasive weed along with roadside, agricultural fields or/and in baron lands of Rajasthan, India, therefore, it may act as a reservoir host for AEV and ALCB, and become a threat to cultivation of sunflower or other commercially cultivated plats. Moreover, the first report of AEV and ALCB betasatellite on a new weed host i.e. wild sunflower (Helianthus spp.) is a novelty.
Acknowledgement: Authors are thankful to Director, CSIRNBRI, Lucknow for facilities. This study was partially funded by the Council of Scientific and Industrial Research, New Delhi, India (Grant number BSC-0117).
References
- Sujatha M. WILD Helianthus species used for broadening the genetic base of cultivated sunflower in India. Helia. 2006;29:77-6.
- Vanitha LS. Molecular characterization of Begomovirus causing leaf curl disease in sunflower. Ph.D. Thesis. University of Agricultural Sciences, Bengaluru, India. 2012.
- Moffat AS.Gemini viruses emerge as serious crop threat. Science.1999;286:1835.
- Lapidot M, Gelbart D, Gal-On A, et al. Frequent migration of introduced cucurbit-infecting Begomoviruses among Middle Eastern countries. Virology J.2014;11:181.
- Fauquet CM, Bisaro DM, Briddon RW, et al.Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of Begomovirus species. Arch Virol.2003;148:405-1.
- Briddon RW, Mansoor S, Bedford ID, et al. Clones of cotton leaf curl Geminivirus induce symptoms atypical of cotton leaf curl disease. Virus Genes.2000;20:17-4.
- Briddon RW, Bull SE, Amin I, et al. Diversity of DNA betasatellite molecule associated with some monopartite Begomoviruses. Virology.2003;312:106-1.
- Huang C, Xie Y, Zhao L, et al. A naturally occurring defective DNA satellite associated with a monopartite Begomovirus: Evidence for recombination between alpha satellite and betasatellite. Viruses.2013;5:2116-8.
- Briddon RW, Brown JK, Moriones E, et al. Recommendations for the classification and nomenclature of the DNA-β satellites of Begomoviruses. Arch Virol.2008; 153:763-1.
- Zhou X. Advances in understanding Begomovirus satellites. Annu Rev Phytopathol.2013;51:357-1.
- Briddon RW, Bull SE, Amin I, et al.Diversity of DNA 1: A satellite-like molecule associated with monopartite Begomovirus-DNA b complexes. Virology.2004;324:462-4.
- Paprotka T, Metzler V, Jeske H. The first DNA 1-like a satellites in association with New World Begomoviruses in natural infections. Virology.2010;404:148-7.
- Nawaz-Ul-Rehman MS, Nahid N, Mansoor S, et al. Post-transcriptional gene silencing suppressoractivity of two non-pathogenic alphasatellites associated with a Begomovirus. Virology.2010;405:300-8.
- Raj SK, Snehi SK, Khan MS, et al.First molecular identification of a Begomovirus associated with yellow vein net disease in grain amaranth (Amaranthus cruentus L.) in India. Australas Plant Dis Notes.2008;3:129-1.
- Rojas MR, Gilbertson RL, Russel DR, et al. Use of degenerate primers in the polymerase chain reaction to detect whitefly transmitted Geminiviruses. Plant Dis.1993;77:340-7.
- Padidam M, Beachy RN, Fauagequet CM. Tomato leaf curl Geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. J Gen Virol.1995;76:25-5.
- Tamura K, Stecher G, Peterson D, et al. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol.2013;30:2725-9.
- Kumar Y, Hallan V, Zaidi AA. First report of Ageratum enation virus infecting Crassocephalum crepidioides (Benth.) S. Moore and Ageratum conyzoides L. in India. J Gen Plant Pathol.2011;77:214-6.
- Li Z, Xie Y, Zhou X. Tobacco curly shoot virus DNAβ Is Not Necessary for Infection but intensifies symptoms in a host-dependent manner. Phytopathology.2005;95:902-8.
- Raj SK, Snehi SK, Khan M, et al. Detection of Ageratum enation virus from cat’s whiskers (Cleome gynandra L.) with leaf curl symptoms in India. J Gen Plant Pathol.2010;76:292-4.
- Kumar Y, Bhardwaj P, Hallan V, et al. Detection and characterization of Ageratum enation virus and a nanovirus-like satellite DNA1 from zinnia causing leaf curl symptoms in India. J Gen Plant Pathol.2010;76:395-8.
- Raj SK, Snehi SK, Tiwari AK, et al. First molecular characterization of Ageratum enation virus associated with mosaic disease of pointed gourd (Trichosanthes dioicaRoxb.) in India. Phytoparasitica.2011;39:497.
- Kumar J, Gunapati S, Singh SP, et al. Molecular characterization, and pathogenicity of a carrot (Daucus carota) infecting Begomovirus and associated betasatellite from India. Virus Res.2013;178:478-5.
- Swarnalatha P, Mamatha M, Manasa M, et al. Molecular identification of Ageratum enation virus (AEV) associated with leaf curl disease of tomato (Solanum lycopersicum) in India. Australas Plant Dis Notes.2013;8:67-1.
- Tahir M, Amin I, Haider MS, et al.Ageratum enation virus: A Begomovirus of weeds with the potential to infect crops. Viruses.2015;7:647-5.
- Marwal A, Sahu A, Choudhary D, et al. Complete nucleotide sequence of a Begomovirus associated with satellites molecules infecting a new host Tagetes patula in India. Virus Genes.2013;47:194-8.
- Mubin M, Shahid MS, Tahir MN, et al. Characterization of Begomovirus components from a weed suggests that Begomoviruses may associate with multiple distinct DNA satellites. Virus Genes.2010;40:452-7.
- Mansoor S, Briddon RW, Bull SE, et al. Cotton leaf curl disease is associated with multiple monopartite Begomoviruses supported by single DNA b. Arch Virol.2003;148:1969-6.
- Briddon RW, Stanley J. Sub-viral agents associated with plant-infecting single-stranded DNA viruses. Virology.2006;344:198-10.
- Saunders K, Briddon RW, Stanley J. Replication promiscuity of DNA-β satellites associated with monopartite Begomoviruses; Deletion mutagenesis of the Ageratum yellow vein virus DNA betasatellite localises sequences involved in replication. J Gen Virol.2008;89:3165-2.
- Nawaz-Ul-Rehman MS, Mansoor S, Briddon RW, et al. Maintenance of an old world betasatellite by a new world helper Begomovirus and possible rapid adaptation of the betasatellite. J Virol.2009;83:9347-5.