Research Article - Asian Journal of Biomedical and Pharmaceutical Sciences (2018) Volume 8, Issue 64
Flavonoids from Dicliptera chinensis (L.) Nees Grown in Vietnam and their Anti-Inflammatory Activities
Loi Vu Duc*, Tung Bui Thanh and Tung Nguyen HuuSchool of Medicine and Pharmacy, Vietnam National University, 144 Xuan Thuy Str., Cau Giay, Hanoi, Vietnam
- *Corresponding Author:
- Loi Vu Duc
School of Medicine and Pharmacy
Vietnam National University, Vietnam
Phone: (+84)989313325
Email: ducloi.smpvnu@gmail.com
Accepted on January 23 2018
Keywords
Kaempferol-3-O-β-D-glucopyranoside; Nicotiflorin; Kaempferol-3-O-α-L-rhamnopyranoside-7-β-Dglucopyranoside; Catechin; Quercetin
Introduction
The leaves of Dicliptera chinensis (L.) Nees (Acanthaceae) is an ethnomedicine, which has been commonly used for treatment of inflammatory in folk [1]. Phytochemical studies of this plant demonstrated the presence of glycosides, flavonoids, monoterpenoids: diclipariside A, diclipariside B, diclipariside C, acid vanillic, β–sitosterol, 2,5–Dimethoxy–bezoquinone, daucosterol, lugrandoside and poliumonside [2,3]. Gao Yu-tao has isolated thirteen compounds from petroleum ester extract fraction of Dicliptera chinensis, these were hexatriacontanol, stearic acid, lupenone, lupeol, 4-sitost-4-en-3-one, stigmast-5- en-7-oxo-3β-yl palmitate, β-sitosterol, oleanolic acid, 3β,6β- stigmast-4-en-3,6-diol, 6β-hydroxy-stigmast-4-en-3-one, 3β- hydroxy-stigmast-5-en-7-one, dehydrovomifoliol, and vomifoliol [4]. Previous studies reported that D. chinensis had pronounced bioactivities, including antioxidant, antiinflammatory activities. Gao Ya has showed D. chinensis polysaccharide was effective for liver injury induced by antituberculosis drug, and the mechanism may be associated with its anti-inflammatory action [5]. Other study demonstrated that functional components including flavonoids, polysaccharides and polyphenols from D. chinensis had strong free radical scavenging capacity [6]. Although D. chinensis is used for clinical treatment, there have been very few studies on this plant. Therefore, this paper reports on the phytochemical investigation of D. chinensis and on the evaluation of the antiinflammatory activities of isolated compounds.
Materials and Methods
Plant material
The whole plants of D. chinensis (L.) Nees were collected from Nam Dinh province, Vietnam during June 2016 and taxonomically identified by the Institute of Ecology and Biological Resources, Vietnam Academy of Science and Technology. The samples were stored at the School of Medicine and Pharmacy-VNU.
General experimental procedures
Column chromatography was performed on silica gel (0.040-0.063 mm, Nicalai Tesque Inc., Japan), YMC ODS-A (50 μm, YMC Co. Ltd., Japan).Organic solvents were of analytical grade.Analytical TLC was performed on Kieselgel 60 F254 (Merck) plates (silica gel, 0.25 mm layer thickness) and RP-18 F254 (Merck) plates (0.25 mm layer thickness). Spots were visualized using ultraviolet radiation (at 254 and 365 nm) and by spraying with 10% H2SO4, followed by heating with a heat gun.The NMR [1H (500 MHz), 13C (125 MHz), and DEPT-90 and 135 MHz)] spectra were recorded on an AVANCE spectrometer AV 500 (Brucker, Germany) in the Institute of Chemistry, Vietnam Academy of Science and Technology (VAST).
Spectra (ESI-MS) were recorded on an AGILENT 1260 Series LC-MS ion Trap (Agilent Technologies, USA).Melting points were measured on SMP10 BioCote in the School of Medicine and Pharmacy-VNU.Optical rotation was measured on PLR-4, MRC scientific instruments in the School of Medicine and Pharmacy -VNU.
Extraction and isolation
General experimental procedures were performed as previous study with some modifications [7]. Leaves of D. chinensis was dried, powdered and then extracted with 96% ethanol (8L × 3 h × 3 times) by supersonic method. The resulting extracts were combined and then evaporated to dryness in vacuo to yield crude extract (630.0 g). The ethanol extract (120 g) was dissolved in water (1.2 L) and subjected to liquid-liquid partitioning (3 times) using n-hexane, ethyl acetate (EtOAc), yielding 31.0 g and 56.0 g of residue, respectively. The aqueous fraction was concentrated to yield 33.0 g of residue.
The EtOAc residue (50 g) was separated by column chromatography on silica gel 60, eluting with chloroform/ methanol (30/1 → 1/1, v/v) to obtain 4 fractions (E1 → E4). The E2 fraction (14.2 g) was submitted to chromatography on silica gel 60, eluted with chloroform/acetone/water (2/5/0.1, v/v), yielding 4 sub-fractions (E2.1 → E2.4). Fraction E2.1 (1.9 g) was further applied to an RP-18 column eluting with Acetone/H2O (2:3, v:v), to yield 1 (56 mg). Fraction E2.3 was crystallized on solvents n-hexane/ethyl acetate (3/5, v/v) to yield 2 (54 mg).The aqueous residue (30 g) was subjected to a Diaion HP-20 column, then eluted with 25%, 50%, 75%, and 100% aqueous methanol, yielding 4 fractions (N1 → N4). Fraction N3 (10.0 g) was further separated over a silica gel column and eluted with chloroform/methanol (30/1 → 1/1, v/v) to yield four sub-fractions (N3.1 → N3.4). Fraction N3.1 (6:1:0.1, ethyl acetate: methanol: water, v/v, 1.2 g) was chromatographed on a silica gel 60 column, yielding 3 (36 mg). Fraction N3.2 (2.1 g) was applied to an RP-18 column eluting with methanol/H2O (2/1, v:v) to yield 4 (51 mg). Finally, fraction N3.4 (2.2 g) was purified by silica gel column, and eluted with chloroform/methanol (30:1, v/v) to yield 5 (46 mg).
COX-2 assay
COX-2 assays were performed as described by Zsofia Kutil [8]. Human recombinant COX-2 (0.5 unit/reaction) was added to 180 μL of incubation mixture consisting of 100 mM tris buffer (pH 8), 5 μM porcine hematin, 18 mM L-epinephrine, and 50 μM Na2EDTA. The test substance dissolved in DMSO or pure DMSO (in case of blank) was added (10 μL) and the mixture was preincubated for 5 min at room temperature. The addition of 5 μL of 10 μM arachidonic acid started the reaction. After 20 min incubation at 37°C the reaction was stopped by 10 μL of 10% formic acid. All samples were diluted 1:15 in assay buffer and the concentration of PGE2 produced by the reaction was determined by a PGE2 ELISA kit (Enzo Life Sciences, USA) according to the manufacturer’s instructions. Absorbance relative to PGE2 concentration was measured by microplate reader Tecan Infinite M200 (Tecan Group, Switzerland) at 405 nm. The results were expressed as percentage inhibition of PGE2 formation against untreated samples (blanks). The test was performed in three independent experiments with three replicates.
The percent inhibition was calculated manually using the following equation:

The effects of compounds were expressed by IC50 values. IC50 is defined as the concentration of inhibitor that reduces enzyme activity by 50%. All samples were tested three times to reduce the error and establish a stable baseline.
Results and discussion
Chemical structure elucidation
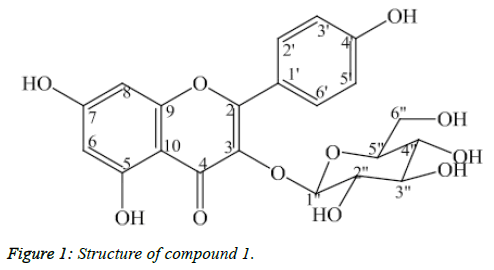
Compound 1: Kaempferol-3-O-β-D-glucopyranoside
Compound 1 was isolated as light yellow powder (Figure 1). The APCI-MS data showed the protonated peak [M+H]+ at m/z 449.0, which matched with the molecular formula of 1 as C21H20O11. The 1H-NMR spectrum showed two doublets at δH 6.23 (1H, d, J=2.0 Hz, H6) and δH 6.42 (1H, d, J=2.0 Hz, H8) corresponding to C6- and C8–protons of the flavonol A ring. Two doublets at δH 6.91 (2H, d, J=9.0 Hz, H3’, H5’) and δH 8.07 (2H, d, J=9.0 Hz, H2’, H6’) corresponded to their parasubstituted flavonoid B-ring. The anomer proton signal at δH 5.26 (1H, d, J=7.5 Hz, H1’) suggested that 1 was a flavonoid glycoside.
The 13C-NMR and DEPT confirmed the presence of 21 carbon atoms. Besides the 15 carbon signals of the flavonoid nucleus, the 13C-NMR spectrum of 1 exhibited six carbon resonances of a sugar moiety. In addition, the HMBC correlation between the anomeric proton δH 5.26 (H1”) and the carbon signal δC 135.47 (C3) revealed the linkage with the aglycone moiety which is shown in Table 1. Based on above deductions and comparing spectral data to reference [9].1 was determined as kaempferol-3- O-β-D-glucopyranoside.
| Position | #δC | δC | DEPT | δH (J=Hz) | HMBC (H→C) |
|---|---|---|---|---|---|
| 2 | 159.14 | 159.07 | C | - | |
| 3 | 135.46 | 135.47 | C | - | |
| 4 | 179.53 | 179.51 | C | - | |
| 5 | 163.04 | 163.09 | C | - | |
| 6 | 99.93 | 100.03 | CH | 6.23 (d, 2.0) | 5,8 |
| 7 | 165.99 | 166.42 | C | - | |
| 8 | 94.79 | 94.85 | CH | 6.42 (d, 2.0) | 7, 9, 10 |
| 9 | 158.51 | 158.57 | C | - | |
| 10 | 105.74 | 105.65 | C | - | |
| 1' | 122.8 | 122.83 | C | - | |
| 2' | 132.28 | 132.27 | CH | 8.07 (d, 9,0) | |
| 3' | 116.1 | 116.09 | CH | 6.91 (d, 9,0) | 1' |
| 4' | 161.54 | 161.58 | C | - | |
| 5' | 116.1 | 116.09 | CH | 6.91 (d, 9,0) | |
| 6' | 132.28 | 132.27 | CH | 8.07 (d, 9,0) | 5', 4' |
| 1'' | 104.09 | 104.14 | CH | 5.26 (d,7,5) | |
| 2'' | 75.73 | 75.75 | CH | 3.45 (m) | 3 |
| 3'' | 78.42 | 78.42 | CH | 3.24 (m) | |
| 4'' | 71.35 | 71.38 | CH | 3.37 (m) | |
| 5'' | 78.38 | 78.07 | CH | 3.45 (m) | |
| 6'' | 62.61 | 62.65 | CH2 | 3.71 (dd, 2.0, 11.5) 3.52 (dd, 5.5, 12.0) |
Table 1: MR spectral data for compound 1, #: 13C-NMR data of kaempferol-3-0-β-D-glucopyranoside in CD30D [12].
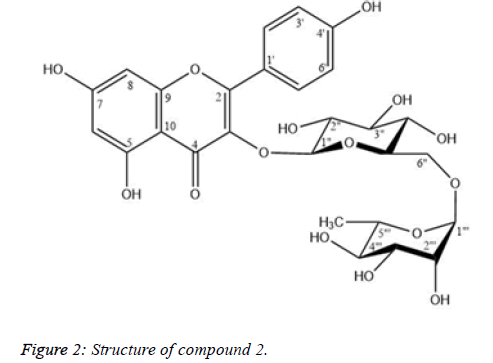
Compound 2: Nicotiflorin
Yellow needless.
Melting point: 190-192 oC.
[α]D25=+20.2 (c = 0.5, MeOH);
Molecula fomula: C27H30O15 ; M=594;
1H-NMR and 13C-NMR (DMSO-d6) data: Table 2.
| Position | #δC | δC | DEPT | δH (J=Hz) | HMBC (H→C) |
|---|---|---|---|---|---|
| 2 | 158.5 | 156.89 | C | - | - |
| 3 | 135.5 | 133.27 | C | - | - |
| 4 | 179.4 | 177.42 | C | - | - |
| 5 | 163 | 161.23 | C | - | - |
| 6 | 100 | 98.79 | CH | 6.20 (d, 2.0) | 5, 7, 8, 10 |
| 7 | 166.1 | 164.26 | C | - | |
| 8 | 94.9 | 93.8 | CH | 6.40 (d, 2.0) | 6, 7, 9, 10 |
| 9 | 159.4 | 156.55 | C | - | - |
| 10 | 105.6 | 104 | C | - | - |
| 1′ | 122.7 | 120.94 | C | - | - |
| 2′, 6′ | 132.4 | 130.9 | CH | 7.98 (d, 8.5) | 2, 2′, 3′, 4′, 5′, 6′ |
| 3′, 5′ | 116.1 | 115.14 | CH | 6.88 (d, 9.0) | 1′, 2′, 3′, 4′, 5′, 6′ |
| 4′ | 161.5 | 159.92 | C | - | - |
| 1′′ | 104.6 | 101.4 | CH | 5.31 (d, 7.5) | 3, 3′′ |
| 2′′ | 75.7 | 74.22 | CH | 3.16 * | 1′′ |
| 3′′ | 78.1 | 75.78 | CH | 3.24* | 2′′ |
| 4′′ | 71.4 | 69.98 | CH | 3.06* | 3′′, 6′′ |
| 5′′ | 77.2 | 76.42 | CH | 3.22 (m) | 6′′ |
| 6′′ | 68.5 | 66.93 | CH2 | 3.27* | |
| 3.69 (d 10.5) | 4′′ | ||||
| 1′′′ | 102.4 | 100.79 | CH | 4.38 (br s) | 6 ′′, 2′′′, 3′′′, 5′′′ |
| 2′′′ | 72.1 | 70.38 | CH | 3.42* | 3′′′, 4′′′ |
| 3′′′ | 72.3 | 70.65 | CH | 3.28* | 2′′′, 4′′′ |
| 4′′′ | 73.9 | 71.88 | CH | 3.09* | 2′′′, 3′′′, 6′′′ |
| 5′′′ | 69.7 | 68.27 | CH | 3.27* | 3′′′ |
| 6′′′ | 17.9 | 17.72 | CH3 | 0.98 (d, 6.0) | 4′′′, 5′′′ |
Table 2: NMR spectral data for compound 2, #13C-NMR data of nicotiflorin [8], :*overlap.
Compound 2 was isolated as yellow needless (Figure 2). The 1H and 13C-NMR spectral data of 2 were similar to those of 1, except for the presence of a rhamnose unit and the strong downfield shift of a methylene carbon at δC 66.93 (C-6′′). The (1 → 6) glycosidic bond, of rhamnose to glucose was characterized from the cross-peak of H-1‴ (δH 4.38) to C-6″ (δC 66.93) and the downfield shift of C-6″ of the glucose unit which is shown in Table 2. By comparing physicochemical properties and spectroscopic data in the literature [10,11], 2 was identified as nicotiflorin.
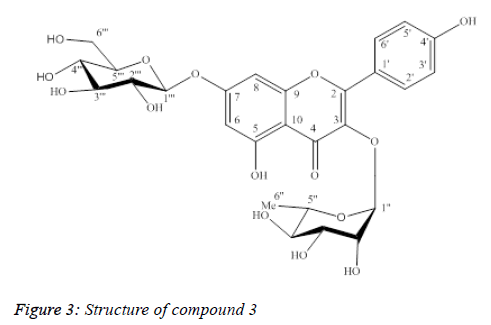
Compound 3: Kaempferol-3-O-α-L-rhamnopyranoside-7-β-Dglucopyranoside
Compound 3 was isolated as yellow powder (Figure 3).
UV λmax (MeOH): 265 nm, 344 nm.
The ESI-MS data showed in Table 3, the protonated peak [M +H]+ at m/z 595.1, which matched with the molecular formula of 3 as C27H30O15. The 1H-NMR spectrum of 3 showed metacoupled aromatic protons (A-ring) at δH 6.51 (1H, d, J=2.0 Hz, H6) and δH 6.76 (1H, d, J=2.0 Hz, H8); ortho-coupled aromatic protons (B-ring) at δH 7.82 (2H, d, J=9.0 Hz, H2’, H6’) and δH 6.97 (2H, d, J=8.0 Hz, H3’, H5’), suggesting the structure of kaempferol aglycone to 3. The presence of rhamnose and glucose moieties was also suggested by two anomer proton signals at δH 5.42 (1H, d, J=1.5Hz, H1”) and δH 5.09 (1H, d, J=7.0 Hz, H1”’), one methyl group at δH 0.96 (3H, d, J=6.0 Hz, H6”), one oxymethilene group at δH 3.74 (1H, m, H6”’α), δH 3.97 (1H, dd, J=12.0/ 2.0 Hz, H6”’β).
| Position | #δC | δC | DEPT | δH Mult., (J inHz) | HMBC (H → C) |
|---|---|---|---|---|---|
| 2 | 159.7 | 159.85 | C | - | |
| 3 | 136.4 | 136.46 | C | - | |
| 4 | 179.7 | 179.8 | C | - | |
| 5 | 162.7 | 162.8 | C | - | |
| 6 | 100.8 | 100.86 | CH | 6.51 (d, 2.0) | 5,7,8,10 |
| 7 | 164.6 | 164.66 | C | ||
| 8 | 95.7 | 95.82 | CH | 6.76 (d, 2.0) | 6,7,9,10 |
| 9 | 158 | 158 | C | ||
| 10 | 107.6 | 107.69 | C | ||
| 1’ | 122.3 | 122.42 | C | ||
| 2’ | 132 | 132.02 | CH | 7.82 (d, 9.0) | 2,4’ |
| 3’ | 116.5 | 116.57 | CH | 6.97 (d, 8.0) | 1’,4’ |
| 4’ | 161.7 | 161.75 | C | ||
| 5’ | 116.5 | 116.57 | CH | 6.97 (d, 8.0) | 1’,4’ |
| 6’ | 132 | 132.02 | CH | 7.82 (d, 9.0) | 2,4’ |
| 1’’ | 103.4 | 103.48 | CH | 5.42 (d, 1.5) | 3, 5’’ |
| 2’’ | 71.2 | 71.27 | CH | 3.36 (m) | |
| 3’’ | 72.1 | 72.07 | CH | 3.74 (m) | |
| 4’’ | 73.1 | 73.17 | CH | 3.36 (m) | |
| 5’’ | 71.8 | 71.89 | CH | 3.74 (m) | |
| 6’’ | 17.6 | 17.64 | CH3 | 0.96 (d, 6,0) | 4’’, 5’’ |
| 1”’ | 101.5 | 101.58 | CH | 5.09 (d, 7,0) | 7 |
| 2’’’ | 75.1 | 74.71 | CH | 3.52 (m) | |
| 3’’’ | 78 | 77.81 | CH | 3.54 (m) | |
| 4’’’ | 72.1 | 72.12 | CH | 3.36 (m) | |
| 5’’’ | 78.3 | 78.36 | CH | 3.54 (m) | |
| 6’’’ | 62.4 | 62.46 | CH2 | 3.74 (m)3.97 (dd, 12.0, 2.0) |
Table 3: NMR spectral data for compound 3,#: 13C-NMR data of kaempferol-3-0-α-L-rhamnopyranoside-7-β-D-glucopyranoside in CD3OD[5]
The 13C-NMR and DEPT spectra showed 27 carbon resonances for 3,15 of which were identical to kaempferol. The 1H and 13C NMR spectral data together with ESI-MS supported the presence of two hexose units in 3. A HMBC experiment performed with 3, showed correlations between the anomeric proton at δH 5.42 (1H, d, J=1.5 Hz, H1”) and the quaternary carbon atom at δC 136.46 (C3); the proton at δH 5.09 (1H, d, J=7.0 Hz, H1”’) and the carbon atom at δC 164.66 (C7), indicating the sites of glycosidation. The NMR data of 3 showed strong agreement with those of kaempferol 3-O-α-Lrhamnopyranoside- 7-O-β-D-glucopyranoside [12]. By comparison with previously reported literature, the structure of 3 was deduced as kaempferol 3-O-α-L- rhamnopyranoside-7- O-β-D-glucopyranoside.
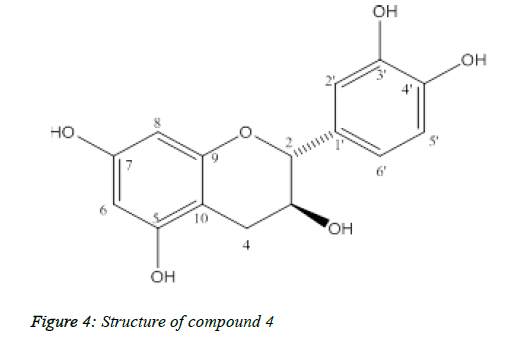
Compound 4: (+)-catechin
Compound 4 was isolated as yellow powder, melting point: 175-177°C (Figure 4).
The ESI-MS spectral showed molecular ion peak at m/z 291 [M+H]+ and 289 [M-H]-, consistent with the molecular formula C15H14O6. The 13C-NMR of 4 showed 15 carbon signals including twelve aromatic carbons, two oxygenated aliphatic carbons at δC 82.82 and 68.79 and one aliphatic carbon at δC 28.42. The 1H-NMR indicated in Table 4, flavan 3-ol moiety including 1,3,4-substituted aromatic proton signals at δH 6.86 (1H, d, J=2.0, H-2'), 6.78 (1H, d, J=8.5, H-5'), 6.75 (1H, dd, J=8.5, 2.0, H-6'), two meta-coupled aromatic proton signals at δH 5.95 (1H, d, J = 2.5, H-6), 5.89 (1H, d, J=2.5, H-8) and a methene group at δH 2.53 (1H, dd, J=16.0, 8.0, H-4α), 2.87 (1H, dd, J=16.0, 5.5, H-4β) and two methine protons at δH 4.59 (1H, d, J=7.5, H-2), 4.01 (1H, m, H-3). The 2,3-trans configuration was confirmed from the large J value of H-2. Based on the spectroscopic evidences and comparison with literature values, 4 was determined to be (+)-catechin.
| Position | δC [3] | δC | DEPT | δH Mult., (J inHz) | HMBC (H → C) |
|---|---|---|---|---|---|
| 2 | 82 | 82.82 | CH | 4.59 d (7.5) | 3, 4, 9, 1′, 2′, 6′ |
| 3 | 67.9 | 68.79 | CH | 4.01 m | 2, 10, 1′ |
| 4 | 28.1 | 28.42 | CH2 | 2.53 dd (8.0, 16.0) | |
| 2.87 dd (5.5, 16.0) | |||||
| 2, 3, 5, 10 | |||||
| 5 | 156.3 | 156.89 | C | ||
| 6 | 96.7 | 96.42 | CH | 5.95 d (2.5) | 5, 7, 8, 10 |
| 7 | 156.9 | 157.79 | C | ||
| 8 | 95.7 | 95.62 | CH | 5.89 d (2.5) | 6, 7, 9, 10 |
| 9 | 156.7 | 157.51 | C | ||
| 10 | 100.9 | 100.91 | C | ||
| 1′ | 131.5 | 132.27 | C | ||
| 2′ | 115.6 | 115.3 | CH | 6.86 d (2.0) | 3′, 6′ |
| 3′ | 145.4 | 146.18 | C | ||
| 4′ | 145.5 | 146.2 | C | ||
| 5′ | 116.6 | 116.14 | CH | 6.78 d (8.5) | 3′, 4′ |
| 6′ | 120.3 | 120.05 | CH | 6.75 dd (2.0, 8.5) | 4′ |
Table 4: NMR spectral data for compound 4, #: 13C-NMR data of (+)- catechin [13]
Compound 5: Quercetin
Yellow needless. Melting point: 313-314 °C.
Molecula fomula: C15H10O7 ; M= 302.
1H-NMR and 13C-NMR (CD3OD) data: Table 5
| Position | #δC | δC | δH (J=Hz) |
|---|---|---|---|
| 2 | 146.8 | 147.72 | |
| 3 | 135.6 | 137.19 | |
| 4 | 175.7 | 177.27 | |
| 5 | 160.6 | 162.45 | |
| 6 | 98.1 | 99.22 | 6.19 (d, 2.0) |
| 7 | 163.8 | 165.51 | |
| 8 | 93.3 | 94.4 | 6.39 (d, 2.0) |
| 9 | 156.1 | 158.19 | |
| 10 | 103 | 104.49 | |
| 1′ | 121.9 | 124.13 | |
| 2′ | 115.1 | 115.98 | 7.74 (d, 2.0) |
| 3′ | 145 | 146.17 | |
| 4′ | 147.6 | 148.72 | |
| 5′ | 115.5 | 116.2 | 6.89 (d, 8.5) |
| 6′ | 119.9 | 121.67 | 7.64 (dd, 2.0, 8.5) |
Table 5: NMR spectral data for compound 5, #: 13C-NMR dat of quercetin [14]
Compound 5 was obtained as yellow needless (Figure 5). The 1H-NMR spectrum of 5 showed 1,3,4-substituted aromatic proton signals at δH 7.74 (1H, d, J=2.0, H-2'), 6.89 (1H, d, J=8.5, H-5'), 7.64 (1H, dd, J=8.5, 2.0, H-6'), two meta-coupled aromatic proton signals at δH 6.19 (1H, d, J=2.0, H-6), 6.39 (1H, d, J=2.0, H-8). The 13C-NMR spectrum indicated that 5 has a flavonol skeleton with 15 carbons, including five aromatic CH; ten quaternary carbons (one carbonyl, five Obearing, and four aliphatic), suggesting that it is 3,5,7,3′,4′- pentahydroxyflavone, commonly known as quercetin [14].
Anti-inflammatory activities of isolated compounds
Table 6: Inhibitory effect of isolated compunds on COX-2 enzyme Cyclooxygenase enzyme (COX, prostaglandin endoperoxide synthase) catalyzes two reactions, the first reaction adds an atom of oxygen to arachidonic acid to form prostaglandin G2 (PGG2) and the second reaction catalyzes the conversion of PGG2 to PGH2. Thus, the COX enzyme performs an important step in converting arachidonic acid into proinflammatory elements, including prostaglandins, thromboxanes and prostacyclins. Prostaglandins regulate smooth muscle contraction, blood pressure and platelet aggregation, regulate pain and fever. The nonsteroidal anti-inflammatory drugs (NSAIDs) inhibit activity of cyclooxygenase enzyme and therefore it is used for the treatment of analgesic, antipyretic,anti-inflammatory and anti-thrombotic [15]. COX-1 isoform is responsible for regulating activity of the physiological prostanoids in the body. In contrast, COX-2 is an isoform that is involved in inflammatory processes[16]. Long-term administration and uncontrolled NSAIDs cause some serious side effects, such as intestinal bleeding due to inhibiting COX-1 enzyme [15].
| Compound | Concentration (ug/ml) | % Inhibition | IC50 |
|---|---|---|---|
| Kaempferol-3-O-β-D-glucopyranosid | 3.125 | 9.24 ± 1.8 | 38.01 ± 1.89 |
| 6.25 | 15.74 ± 1.2 | ||
| 12.5 | 32.59± 1.2 | ||
| 25 | 40.25 ± 1.5 | ||
| 50 | 56.81 ± 2.7 | ||
| 100 | 86.57 ± 1.2 | ||
| Kaempferol-3-O-α-L-rhamnopyranosid-7-β-D-glucopyranosid | 3.125 | 12.35 ± 2.3 | 22.38 ± 1.72 |
| 6.25 | 19.47 ± 2.1 | ||
| 12.5 | 32.57 ± 2.6 | ||
| 25 | 48.71 ± 2.3 | ||
| 50 | 69.34 ± 1.8 | ||
| 100 | 87.11 ± 1.7 | ||
| Catechin | 3.125 | 14.17 ± 2.7 | 22.95 ± 1.87 |
| 6.25 | 24.29 ± 2.1 | ||
| 12.5 | 36.57 ± 2.7 | ||
| 25 | 52.39 ± 1.2 | ||
| 50 | 69.53 ± 2.4 | ||
| 100 | 87.08 ± 1.8 | ||
| Quercetin | 3.125 | 10.78 ± 1.8 | 37.15 ± 1.35 |
| 6.25 | 19.67 ± 1.2 | ||
| 12.5 | 30.14 ± 1.5 | ||
| 25 | 44.29 ± 2.1 | ||
| 50 | 53.78 ± 1.7 | ||
| 100 | 78.24 ± 1.9 | ||
| Nicotiflorin | 3.125 | 12.36 ± 1.9 | 32.35 ± 1.94 |
| 6.25 | 19.87 ± 2.3 | ||
| 12.5 | 28.34 ± 2.1 | ||
| 25 | 45.89 ± 2.2 | ||
| 50 | 61.27 ± 1.4 | ||
| 100 | 87.24 ± 2.4 | ||
| Celecoxib (positive drug control) |
0.1563 | 12.36 | 1.43 ± 0.12 |
| 0.3125 | 19.87 | ||
| 0.625 | 28.34 | ||
| 1.25 | 45.89 | ||
| 2.5 | 61.27 | ||
| 5 | 87.24 | ||
| 10 | 98.25 |
Table 6: Inhibitory effect of isolated compunds on COX-2 enzyme
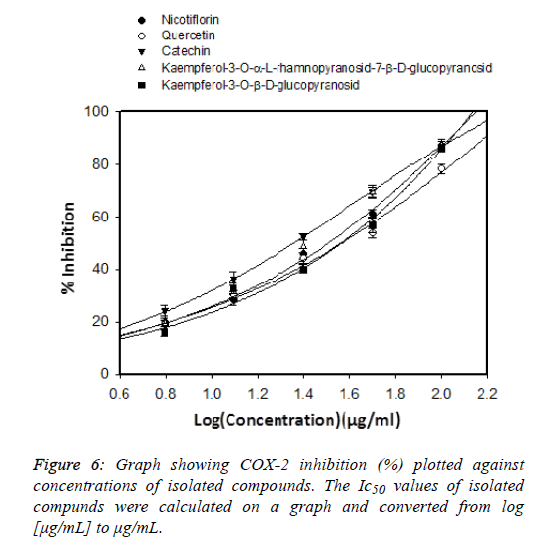
New-generation COX-2 inhibitors do not induce intestinal bleeding but can cause cardiovascular-related adverse events [17]. Steroid drugs play an important role in the treatment of inflammatory diseases, but it is only used in the short term due to their toxicity. Therefore, finding natural compounds with anti-inflammatory effects but with few side effects is very urgent. Medicinal plants play an important role in the search for potential compounds for drug development. The screening process of natural anti-inflammatory compounds has been increasing in recent years [18]. In this study we evaluated the COX-2 inhibitory effect of the isolated compounds shown in Figure 6. Celecoxib was used as positive control to inhibit COX2 enzyme shown in Figure 7 and its IC50 value was of 1.43 ± 0.12 μg / mL. Previous published showed that the COX-2 inhibitory effect of Celecoxib had an IC50 of 0.34 μg/mL [19]. which is shown in Table 7. This shows that our results are consistent with previous publications. Among five isolated flavonoids, two compounds showed potential COX-2 inhibitory effects, Kaempferol-3-O-α-L-rhamnopyranosid-7-β- D-glucopyranosid (IC50=22.38 ± 1.72 μg/mL), and catechin (IC50=22.95 ± 1.87μg/mL). Previous study showed that kaempferol is a potent strong inhibitor COX-2 which could be chemopreventive agent against cancer [20,21]. In-Ja Par has demonstrated that catechin could decrease the cyclooxygenase-2 (COX-2) expression and prostaglandin E2 (PGE2) levels and reduce the excessive ROS [22]. From the results of this study, we selected catechin and kaempferol-3-O- α-L-rhamnopyranoside-7-β-D-glucopyranoside for further studies of their anti-inflammatory activities.
| Sample | LogIC50 | IC50 |
|---|---|---|
| Kaempferol-3-O-α-L-rhamnopyranosid-7-β-D-glucopyranosid | 1.35 | 22.38 |
| Catechin | 1.36 | 22.95 |
| Quercetin | 1.57 | 37.15 |
| Kaempferol-3-O-β-D-glucopyranosid | 1.58 | 38.01 |
| Nicotiflorin | 1.51 | 32.35 |
Table 7: IC50 of isolated compunds on COX-2 enzyme
Conclusion
From the leaves of D. chinensis (L.) Nees (Acanthaceae) collected in Nam Dinh province five flavonoids (1-5) were isolated by chromatographic methods. On the basis of spectroscopic analyses and by spectral comparison with published literature, the isolated compounds were identified as Kaempferol-3-O-β-D-glucopyranoside (1), Nicotiflorin (2), Kaempferol-3-O-α-L-rhamnopyranoside-7-β-Dglucopyranoside (3), Catechin (4), Quercetin (5). This is the first report on the isolation of 1 and 3 from Dicliptera genus. Among five isolated compounds, 3 and 4 also showed significant COX-2 inhibitory effects, with IC50 values of 22.38 ± 1.72, 22.95 ± 1.87 μg/mL, respectively which is shown in Table 6.
Acknowledgement
The research was supported by has been financed by Vietnam National University, Hanoi with grants number: QG.17.28These authors have declared that there is no conflict of interest.
References
- Lin CC, Lin ML, Lin JM. The antiinflammatory and liver protective effect of Tithonia diversifolia (Hemsl.) gray and Dicliptera chinensis Juss. Extracts in rats. Phytother Res. 1993;7: 305-309.
- Luo Y, Feng , Tian Y, Zhang G. Glycosides from Dicliptera riparia. Phytochem. 2002;61:449-454.
- Zhang X ,Zhang J, Jia L, Xiao S. Dicliptera Chinensis polysaccharides target TGF-β/Smad pathway and inhibit stellate cells activation in rats with dimethyl nitrosamine-induced hepatic fibrosis. Cell and mol biol (Noisy-le-Grand, France). 2016;62:99-103.
- Gao Yt, Yang Xw, Ai Tm. Chemical constituents of whole herb of Dicliptera chinensis. Chin Trad and Herbal Drugs. 2007;38:14.
- Gao Y , Zhong M, Zhong J, Zhang K. Study of Dicliptera Chinensis Polysaccharide in Counteracting Liver Injury Induced by Antituberculosis Drugs. J of Guangzhou University of Trad Chin Med. 2014;6:26.
- Gao Y, Zhong M, Zhong G, Zhang K. Study of Dicliptera Chinensis Polysaccharide in Counteracting Liver Injury Induced by Antituberculosis Drugs. Journal of Guangzhou University of Traditional Chinese Medicine.2014;6:26.
- LI Nw, LIU Ch, LU Yj. Antioxidant Activities of Functional Components from Dicliptera chinensis. Food Sci. 2011;13:18.
- Thanh TB, Duc LV, Thanh HN, Tien VN. In vitro antioxidant and anti-inflammatory activities of isolated compounds of ethanol extract from Sanchezia speciosa Leonard’s leaves. J of basic and clin physiol and pharmacol. 2017;28:79-84.
Kutil Z, Kvasnicova M, Temml Marie V, Schuster D, Marsik P, Cusimamani EF. Isolation and identification of compounds from the ethanolic extract of flowers of the tea (Camellia sinensis) plant and their contribution to the antioxidant capacity. LWT-Food Sci and Technol. 2009; 42: 1439-1443. - Yang Z, Tu Y, Baldermann S, Dong F, Watanabe N.Isolation and identification of compounds from the ethanolic extract of flowers of the tea (Camellia sinensis) plant and their contribution to the antioxidant capacity. LWT-Food Sci and Technol. 2009;42:1439-1443.
- Eldahshan OA, Ayoub NA, Singab AN, Al-Azizi MM. Potential superoxide anion radical scavenging activity of doum palm (Hyphaene thebaica L.) leaves extract. Records of Nat Prod. 2008;2:83.
- Park SY, Lee SY, Kang SS. Chemical constituents of Lathyrus davidii. Nat Prod Sci. 2008;14: 281-288.
- Chen YH, Chen YH, Chang FR, Lin YJ, Chen JF, Wu YC, Wu MJ. Identification of phenolic antioxidants from Sword Brake fern (Pteris ensiformis Burm.). Food chem. 2007;105: 48-56.
- Davis AL, Cai Y, Davies AP, Lewis JR. 1H and 13C NMR assignments of some green tea polyphenols. Magn reson in Chem. 1996;34:887-890.
- Shen CC, Chang YS, Hott LK. Nuclear magnetic resonance studies of 5,7-dihydroxyflavonoids. Phytochem. 1993;34:843-845.
- Lee JL, Mukhtar H, Bickers DR, Kopelovich L, Athar M. Cyclooxygenases in the skin: pharmacological and toxicological implications. Toxicol and appl pharmacol. 2003;192:294-306.
- Vane J. Towards a better aspirin. Nat. 1994;367:215.
- Mukherjee D, Nissen SE, Topol EJ. Risk of cardiovascular events associated with selective COX-2 inhibitors. 2001;286:954-959.
- Chen S. Natural products triggering biological targets-a review of the anti-inflammatory phytochemicals targeting the arachidonic acid pathway in allergy asthma and rheumatoid arthritis. Curr drug targets. 2011;12:288-301.
- Botting R. Inhibitors of cyclooxygenases: mechanisms, selectivity and uses. J of Physiol and Pharmacol. 2006;57:113.
- Lee KM, Lee KW, Jung SK, Lee EJ, Heo YS, Bode AM, Lubet RA, Lee HJ, Dong Z. Kaempferol inhibits UVB-induced COX-2 expression by suppressing Src kinase activity. Biochem Pharmacol. 2010; 80:2042-2049.
- García-Mediavilla V, Crespo I, Collado PS, Esteller A, Sánchez-Campos S, Tuñón MJ, González-Gallego J. The anti-inflammatory flavones quercetin and kaempferol cause inhibition of inducible nitric oxide synthase, cyclooxygenase-2 and reactive C-protein, and down-regulation of the nuclear factor kappa B pathway in Chang Liver cells. Eur J of Pharmacol. 2007;557:221-229.
- Park IJ, Lee YK, Hwang JT, Kwon DY, Ha J, Park OJ. Green Tea Catechin Controls Apoptosis in Colon Cancer Cells by Attenuation of H2O2‐Stimulated COX‐2 Expression via the AMPK Signaling Pathway at Low‐Dose H2O2. Ann of the New York Acad of Sci. 2009;1171:538-544.